- BiologyDiscussion.com
- Follow Us On:
- Google Plus
- Publish Now


Denaturation and Renaturation of DNA
ADVERTISEMENTS:
In this article we will discuss about Denaturation and Renaturation of DNA Double Helix.
Denaturation of DNA:
Denaturation of DNA double helix takes place by the following denaturating agents:
(i) Denaturation by Temperature :
If a DNA solution is heated to approximately 90°C or above there will be enough kinetic energy to denature the DNA completely causing it to separate into single strands. This denaturation is very abrupt and is accelerated by chemical reagents like urea and formamide.
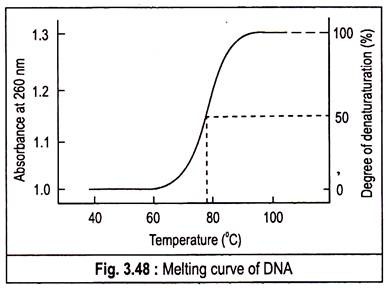
The chemicals enhance the aqueous solubility of the purine and pyrimidine groups. This separation of double helix is called melting as it occurs abruptly at a certain characteristic temperature called denaturation temperature or melting temperature (T m ).
It is defined as temperature at which 50% of the DNA is melted. The abruptness of the transition indicates that the DNA double helix is highly cooperative structure, held together by many reinforcing bonds. The melting of DNA can be followed spectrophotometrically by monitoring the absorbance of DNA at 260 nm. T m is analogous to the melting point of crystal. The T m value depends on the nature of the DNA.
If several samples of DNA are melted, it is found that the T m is highest for those DNAs that contain the highest proportion of G—C. Actually the value is used to estimate the percentage of G—C in a DNA sample. In fact, the T m of DNA from many species varies linearly with G—C content.
This relationship between T m and G—C content arises due to guanine and cytosine form three hydrogen bonds when base paired, whereas adenine and thymine form only two.
Denaturation involves the following changes of the properties of DNA:
(a) Increase in Absorption of UV-Light:
If denaturation is followed spectrophotometrically by monitoring the absorbance of light at 260 nm, it is observed that the absorbance at 260 nm increases as the DNA become denatured, a phenomenon known as the hyperchromatic effect or hyperchromacity or hyperchromism. This is due to un-stacking of base pairs.
A plot of the absorbance at 260 nm against the temperature of a DNA solution indicates that little denaturation occurs below approximately 70°C, but further increases in temperature result in a marked increase in the extent of denaturation.
(b) Decrease in Specific Optical Rotation:
Double-stranded DNA shows a strong positive rotation which highly decreases with denaturation. This change is analogous to the change in rotation observed when the proteins are denatured.
(c) Decrease in Viscosity:
The solutions of native DNA exhibit high viscosity because of the relatively rigid double helical, long and rod like character of DNA molecule. Denaturation causes a marked decrease in viscosity.
If melted DNA is cooled it is possible to reassociate the separated strands, a process known as renaturation. However, a stable double-stranded molecule may be formed only if the complementary strands collide in such a way that their bases are paired precisely. But renaturation may not be precise if the DNA is very long and complex.
Thus the rate of renaturation (renaturation kinetics) can give information about the complexity of a DNA molecule. Complete denaturation is not a readily reversible process. If a heat-denatured DNA solution is cooled slowly (anneling) and hold the solution at about 25°C below T m and above a concentration of 0.4M Na + for several hours, some amount of
DNA (50-60%) is renatured. Rapid cooling does not reverse denaturation, but if the cooled solution is again heated and then cooled slowly, renaturation takes place.
(ii) Denaturation by Chemical Agents :
Denaturation of DNA double helix can also be brought about by certain chemical agents such as urea and formamide. These chemical reagents enhance the aqueous solubility of the purine and pyrimidine groups. The T m value is lowered by the addition of urea. In 8M urea, T m is decreased by nearly 20°C. DNA can be completely denatured by 95% formamide at room temperature only.
(iii) Effect of pH on Denaturation :
Denaturation also occurs at acidic and alkaline solutions in which ionic changes of the purine and pyrimidine bases can occur. In acidic solutions at pH values 2-3 the amino groups bind with protons and the DNA double helix is disrupted. Similarly, in alkaline solutions at pH 12, the enolic hydroxyl groups ionize, thus preventing the keto-amino hydrogen bonding.
Renaturation of DNA:
When preparations of double-stranded DNA are denatured and allowed to renature, the rate of renaturation can give valuable information about the complexity of the DNA if there are repetitive sequences in the DNA, it shows less complexity in comparison to its total length, but the complexity is equal to its total length if all sequences are unique.
The 1kb DNA fragments are denatured by heating above its T m and then renatured at a temperature 10°C below the T m and monitored either by decrease in absorbance at 260 nm (hypochromic effect), or by passing samples at intervals through a column of hydroxylapatite, which retains only double stranded DNAs, and estimating how much of the sample is retained.
The degree of renaturation after a given time depends on C 0 , the concentration of double stranded DNA prior to denaturation, and t, the duration of the renaturation in seconds. The concentration is measured in nucleotides per unit volume. In order to compare the rates of renaturation of different samples of DNA it is usual to measure C 0 and the time taken for renaturation to proceed half way to completion, t 1/2 , and to multiply these values together to give a C 0 t 1/2 value. The larger the C 0 t 1/2 , the greater the complexity of the DNA; hence λ DNA has a far lower C 0 t 1/2 than does human DNA.
if the extent of renaturation is plotted against log C 0 t (known as Cot curve), it is observed that part of the DNA is renatured quite rapidly while the rest is very slow to renature. This indicates that some sequences have a higher concentration than others i.e., part of the genome consists of repetitive sequences.
These repetitive sequences can be separated from the single-copy unique DNA by passing the renaturating sample through a hydroxylapatite column early in the renaturation process, at a time which gives a low value of C 0 t. At this stage only the rapidly renaturating sequences will be double stranded, and will, therefore, bind to the column.

Related Articles:
- Repeated Sequence of Chromosomal DNA | Genetics
- Physico-Chemical Properties of Nucleic Acids
- Anybody can ask a question
- Anybody can answer
- The best answers are voted up and rise to the top

Forum Categories
- Animal Kingdom
- Biodiversity
- Biological Classification
- Biology An Introduction 11
- Biology An Introduction
- Biology in Human Welfare 175
- Biomolecules
- Biotechnology 43
- Body Fluids and Circulation
- Breathing and Exchange of Gases
- Cell- Structure and Function
- Chemical Coordination
- Digestion and Absorption
- Diversity in the Living World 125
- Environmental Issues
- Excretory System
- Flowering Plants
- Food Production
- Genetics and Evolution 110
- Human Health and Diseases
- Human Physiology 242
- Human Reproduction
- Immune System
- Living World
- Locomotion and Movement
- Microbes in Human Welfare
- Mineral Nutrition
- Molecualr Basis of Inheritance
- Neural Coordination
- Organisms and Population
- Photosynthesis
- Plant Growth and Development
- Plant Kingdom
- Plant Physiology 261
- Principles and Processes
- Principles of Inheritance and Variation
- Reproduction 245
- Reproduction in Animals
- Reproduction in Flowering Plants
- Reproduction in Organisms
- Reproductive Health
- Respiration
- Structural Organisation in Animals
- Transport in Plants
- Trending 14
Privacy Overview

DNA renaturation experiments are useful strategies to estimate the complexity of genomic DNA.
Expert verified solution.
DNA renaturation experiments are used to estimate the complexity of genomic DNA by analyzing the kinetics of renaturation. The rate of renaturation is directly proportional to the complexity of the sequences, with simpler sequences renaturing faster than more complex ones. This allows researchers to determine the complexity and repetition frequency of different genomic sequences
Explanation
- DNA renaturation experiments involve shearing DNA isolates to a specific size
- The sheared DNA is then denatured and allowed to renature under controlled conditions
- The rate of renaturation is measured, and this rate is directly related to the complexity of the genomic sequences
- Simpler sequences renature faster than more complex sequences
- By analyzing the renaturation kinetics, one can determine the complexity and repetition frequency of the genomic sequences
- For educators
- English (US)
- English (India)
- English (UK)
- Greek Alphabet
This problem has been solved!
You'll get a detailed solution that helps you learn core concepts.
Question: DNA renaturation experiments are useful strategies to estimate the number of single nucleotide differences between two species. genetic relatedness between two species.proportion of GC content in an organism's genome. size of an organism's genome.

Not the question you’re looking for?
Post any question and get expert help quickly.

IMAGES
COMMENTS
Chromosome Structure and Organelle DNA Learn with flashcards, games, and more — for free. ... DNA renaturation experiments are useful strategies to estimate the. genetic relatedness between species. 1 / 53. 1 / 53. Flashcards; ... DNA renaturation experiments are useful strategies to estimate the.
DNA renaturation experiments are useful strategies to estimate... DNA polymerase. Enzyme that pairs corresponding nucleotides to preexisting DNA chain to synthesize new DNA. Recruiting low fidelity polymerases allows replication to continue when distortions in the DNA template strand stall synthesis by high fidelity polymerases.
Differences in DNA renaturation rates are used to measure the frequency of specific base sequences, to locate specific sequences, and to analyze species of RNA that anneal to DNA. One important application is in the hybridization experiments such as Southern blots or Northern blots. ... Such DNA hybridization is useful to estimate the ...
Such DNA hybridization is useful to estimate the evolutionary proximity between two different species. DNAs from phylogenetically close species are able to form hybrid molecules in higher frequency than that from species which are far apart. ... Renaturation strategies in purification methods of NC1 domains. ... For DSC experiments, the van't ...
DNA renaturation experiments are useful strategies to estimate the genetic relatedness between two species. When a circular DNA gets underrotated by the action of cellular enzymes, the DNA is said to exhibit
If melted DNA is cooled it is possible to reassociate the separated strands, a process known as renaturation. However, a stable double-stranded molecule may be formed only if the complementary strands collide in such a way that their bases are paired precisely. But renaturation may not be precise if the DNA is very long and complex.
place on a much longer time scale than experiments with synthetic oligonucleotides or homopoly,mers. Reactions may be initiated by cooling a sample from a tem perature above the melting temperature to the renaturation temperature. An alter nate method for DNA renaturation (7) but not DNA· RNA hybridization involves
DNA renaturation experiments are used to estimate the complexity of genomic DNA by analyzing the kinetics of renaturation. The rate of renaturation is directly proportional to the complexity of the sequences, with simpler sequences renaturing faster than more complex ones.
Question: DNA renaturation experiments are useful strategies to estimate the number of single nucleotide differences between two species. genetic relatedness between two species.proportion of GC content in an organism's genome. size of an organism's genome.
The resulted values of k 1 and k 2 are shown in Table 1 and the plot in Fig. 1.The best fit was obtained in case II (χ 2 =0.83) and the conventional model gave a poor fitting (χ 2 =0.71).The renaturation rate constant (k 1) obtained from conventional model was 1×10 10 mol −1 s −1, which indicated that the renaturation should be an ultra fast process.This contradicts the general observation.