An official website of the United States government
Official websites use .gov A .gov website belongs to an official government organization in the United States.
Secure .gov websites use HTTPS A lock ( Lock Locked padlock icon ) or https:// means you've safely connected to the .gov website. Share sensitive information only on official, secure websites.
- Publications
- Account settings
- Advanced Search
- Journal List


Revisiting the malaria hypothesis: accounting for polygenicity and pleiotropy
Emily r ebel, lawrence h uricchio, dmitri a petrov, elizabeth s egan.
- Author information
- Article notes
- Copyright and License information
Correspondence: [email protected] (E.R. Ebel) and [email protected] (E.S. Egan)
Issue date 2022 Apr.
The malaria hypothesis predicts local, balancing selection of deleterious alleles that confer strong protection from malaria. Three protective variants recently discovered in red cell genes are indeed more common in African than European populations. Still, up to 89% of the heritability of severe malaria is attributed to many genome-wide loci with individually small effects. Recent analyses of hundreds of genome-wide association studies (GWAS) in humans suggest that most functional, polygenic variation is pleiotropic for multiple traits. Interestingly, GWAS alleles and red cell traits associated with small reductions in malaria risk are not enriched in African populations. We propose that other selective and neutral forces, in addition to malaria prevalence, explain the global distribution of most genetic variation impacting malaria risk.
Keywords: malaria, red blood cells, evolution, polygenic, pleiotropy
The malaria hypothesis predicts strong selection on protective variants of large effect
Malaria caused by the parasite Plasmodium falciparum has caused hundreds of millions of deaths over the course of human history. Consequently, even deleterious genetic variants that limit malaria severity are expected to be maintained by balancing selection (see Glossary ) within malaria-endemic populations. This “malaria hypothesis” is supported by geographical patterns of monogenic disorders of red blood cells (RBCs) that confer protection against severe malaria ( Box 1 ). For example, the sickle cell allele of hemoglobin beta (HbS) arose and is maintained in sub-Saharan Africa [ 1 ], where the vast majority of malaria deaths occur [ 2 ]. Sickle cell disease and other RBC disorders ( Box 1 ) provide compelling evidence that malaria has imposed strong selective pressures on human populations throughout the world [ 3 ]. Here we focus on selection in Africa from P. falciparum , the deadliest malaria parasite, although RBC disorders and Plasmodium species are found in tropical regions worldwide.
Box 1. History of the malaria hypothesis.
The malaria hypothesis offers an explanation for the high prevalence of hematologic disease in malaria-endemic areas. It is frequently attributed to the population geneticist JBS Haldane, who in the late 1940s, recognized that the prevalence of thalassemia in the Mediterranean was far higher than could be explained by mutation alone [ 65 ]. Thalassemia is a life-threatening anemia caused by a homozygous deletion of one of the genes encoding hemoglobin. Haldane proposed that the causal allele could be maintained within a population, despite its fitness cost in homozygotes, by offering a fitness benefit to heterozygotes—a phenomenon now known as balancing selection. RBCs of heterozygous carriers of thalassemia are unusually small and resistant to hypotonic solutions, so Haldane reasoned that they might also be resistant to Plasmodium parasites. Thalassemia has since been associated with reduced clinical malaria [ 3 ] and slower P. falciparum growth in vitro [ 15 ], although the full mechanism of protection remains unknown.
Also in the mid-twentieth century, the South African scientist AC Allison compiled extensive epidemiological data for sickle cell trait and malaria in Africa [ 66 ]. Similar to thalassemia, sickle cell disease is a life-threatening anemia caused by a homozygous point mutation in the hemoglobin beta gene. Heterozygous carriers lack serious symptoms, though they may have clinically abnormal RBCs. Allison recorded a much higher prevalence of sickle cell trait in malaria-hyperendemic areas of East Africa. In 1954, he experimentally confirmed his own malaria hypothesis by infecting 30 native Luo men with P. falciparum parasites and charting the course of their disease for forty days [ 67 ]. His study provided strong evidence of the anti-malarial effects of HbS, but it included no record of the informed consent process, if any such process existed [ 68 ]. HbS appears to offer protection via hemoglobin polymerization in low oxygen conditions, leading to parasite growth arrest [ 69 ].
The malaria hypothesis is also generally accepted to explain the prevalence of Hemoglobin C disease, Hemoglobin E disease, Southeast Asian Ovalocytosis, and G6PD deficiency [ 70 – 73 ]. In the first three diseases, the heterozygous state provides a clinical advantage against malaria while the rare homozygous state is strongly deleterious. G6PD deficiency, in contrast, involves several common alleles with complex effects on malaria and metabolism [ 71 ]. Anemia can be induced in G6PD-deficient individuals by fava beans, infection, and other factors, though most carriers have a mild deficiency with no symptoms.
The malaria hypothesis is based on RBC disorders with simple genetic causes ( Box 1 ), but it is sometimes extended to polygenic traits like RBC volume, RBC membrane fragility, and malaria resistance itself [ 4 – 9 ]. Here, we explore whether the simple predictions of the malaria hypothesis can be easily generalized to complex traits . Simultaneous analyses of hundreds of genome-wide association studies (GWAS) have suggested that most functional variants in humans have small effects and are pleiotropic , or associated with more than one phenotype. We hypothesize that complex traits related to malaria may not conform to the malaria hypothesis—even when several large-effect alleles do—because of pleiotropy and selective constraint on genetically correlated traits. Furthermore, because most common variants have a global distribution [ 10 ], we expect that most alleles that impact malaria are present in all human populations ( Box 2 ). An overly narrow focus on the malaria hypothesis may distract from progress on the complex biological pathways that contribute to the pathogenesis of severe malaria.
Box 2. Genetic structure and similarity of all human populations.
Most common genetic variation present in humans today is descended from populations living in Africa 100 to 250 thousand years ago (ka) [ 74 ]. Between 40 and 60 ka, a population carrying a subset of this African diversity expanded throughout the rest of the world. As they migrated, human populations experienced a series of bottlenecks that reduced their genetic diversity relative to their distance from Africa [ 75 ]. Some populations of Homo sapiens mixed with Neanderthals and Denisovans [ 74 ], and all populations experienced mutation, recombination, drift , and selection on their unique genetic backgrounds. Genetic variation is clearly structured across human populations ( Figure I ), primarily by geography and demography [ 76 ].
Human population structure allows the regional ancestry of an individual to be inferred from a sufficiently large panel of genetic markers ( Figure I ). For example, around 1.3 million SNPs are common across Africans but rare or undetected in the rest of the world. Since much of this population structure was created by the loss of SNPs in out-of-Africa bottlenecks, it is unlikely that most of these SNPs are strongly related to malaria.
Furthermore, the majority of common genetic variants in humans are shared among all major populations [ 10 ] ( Figure I ), due to our fairly recent common ancestor. This fact was first demonstrated in 1972 using red blood cell genes [ 77 ], including several associated with Plasmodium. In terms of variance partitioning, a large majority (~94%) of human genetic diversity is found within populations, as opposed to across them [ 75 ]. In fact, if we were to randomly sample two members of different populations, they would have on average only ~5% more alleles that differ than two members of the same population [ 75 ].
Despite sharing many alleles inherited from common ancestors, human populations obviously do exhibit differences in phenotypes and allele frequencies. Understanding the extent of local, balancing selection in shaping these differences requires an appropriate neutral (demographic) model; recognition of environmental factors; and knowledge of genetically correlated traits [ 41 , 78 ].
Figure I. Most common human alleles have a broad geographic distribution.
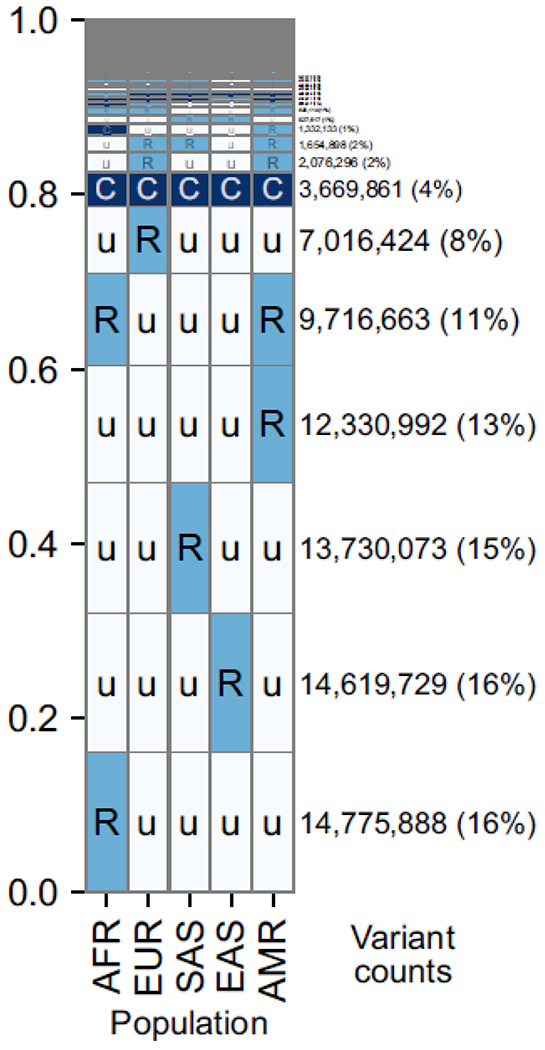
Data are summarized across ~92 million bi-allelic variants from 1000 Genomes. Within each cell, C = common (>5% frequency), R = rare (<5%), u= unobserved (0%). The number and proportion of variants with a given geographical distribution is indicated by the text on the right, as well as the height of the row. AFR=African, EUR=European, SAS=South Asian, EAS=East Asian, AMR=Admixed American. Reproduced from [ 10 ].
Three novel alleles with large protective effects conform to the malaria hypothesis
In the last five years, genetic variants at three RBC loci have been associated with strong protection from severe malaria and replicated in independent studies ( Table 1 ). The first locus, ATP2B4 , encodes the main calcium transporter of RBCs. An ATP2B4 haplotype associated with decreased risk of severe malaria [ 7 ] was recently linked to decreased calcium export and increased dehydration in RBCs [ 7 , 11 – 14 ]. RBC dehydration may directly contribute to malaria resistance by decreasing P. falciparum growth rate in RBCs [ 15 , 16 ]. Similarly, PIEZO1 encodes a mechanosensitive ion channel implicated in xerocytosis, a rare disease characterized by extreme RBC dehydration [ 6 ]. The microsatellite variant PIEZO1 E756del has been associated with strong protection from severe malaria [ 17 ], reduced P. falciparum growth [ 6 , 15 ], and mild RBC dehydration [ 6 , 18 ], although these phenotypes have not been replicated in all studies [ 15 , 17 , 19 , 20 ]. Genes at the glycophorin locus ( GYPA/GYPB ) encode red cell surface receptors coated with sialic acid, which facilitate parasite invasion by specific binding to P. falciparum ligands [ 21 ]. The Dantu allele at this locus encodes a hybrid protein in which the extracellular domain of GYPB is fused to the transmembrane and intracellular domains of GYPA [ 22 ]. Interestingly, the protective effect of Dantu against severe malaria is mediated not through receptor-ligand interactions, but through an increase in RBC membrane tension that inhibits P. falciparum invasion [ 23 ]. For all three of these loci, the same polymorphism that is associated with strong protection from clinical malaria [ 7 , 17 , 22 ] is also associated with RBC trait variation [ 6 , 11 , 14 , 15 , 23 ] and impaired P. falciparum development in vitro [ 6 , 15 , 17 , 23 ].
Novel alleles associated with strong protection from severe malaria.
Based on odds ratio for the broadest category of severe malaria in an additive or recessive association model with one copy of the allele.
mean corpuscular hemoglobin concentration (g/dL red blood cells)
hemoglobin (g/dL whole blood)
red blood cell distribution width (% coefficient of variation)
mean corpuscular volume of red blood cells (fL)
mean cell hemoglobin of red blood cells (pg)
All three protective variants in Table 1 occur at a higher frequency in Africa than Europe, consistent with the malaria hypothesis. PIEZO1 E756del is found in about one third of Africans in 1000 Genomes, which motivated its initial selection for detailed study [ 6 ]. The Dantu variant of GYPA/GYPB has a frequency of 2-13% in some parts of East Africa, but is absent from most of Africa and the rest of the world [ 22 , 24 ]. The protective ATP2B4 haplotype, like variation in the ABO blood group, is found at appreciable frequency (~11-15%) in Europe and South Asia and higher frequency (~35%) across Africa.
The degree of protection observed for these variants in vitro has been smaller than in clinical studies, which has been interpreted as evidence of pleiotropic effects through additional tissues or phenotypes [ 7 , 15 , 17 ]. For example, the protective ATP2B4 haplotype is associated with up to 35% reduction in severe malaria risk, but only ~6% slower growth of P. falciparum in RBCs ( Table 1 ). The same haplotype is associated by GWAS with lymphocyte counts, in addition to RBC dehydration and size [ 25 ], which may contribute to its larger effect on severe malaria. However, part of the difference between GWAS and in vitro models might be explained by limitations of current methods of parasite culture. P. falciparum is typically cultured within RBCs in rich media and at low density, independent of immune cells and normal circulation. These conditions necessarily represent only a fraction of the host biology involved in malaria disease. Furthermore, lab culture conditions are known to select for deleterious parasite traits, including the loss of gametocytogenesis [ 26 ].
Similar to the pleiotropic effects of ATP2B4, PIEZO1 E756del has been linked to macrophage-mediated iron metabolism and athletic jumping performance [ 27 , 28 ] in addition to RBC phenotypes. Both ATP2B4 and PIEZO1 are widely expressed throughout the body, so variants impacting their function could potentially be pleiotropic through immune cells, the vasculature, or other tissue types. Glycophorin proteins are expressed exclusively on RBCs, but the Dantu variant is associated with altered expression of 100 RBC surface proteins [ 23 ] that could be pleiotropic for interactions with immune cells or other issues. The glycophorins and PIEZO1 are both exceptionally polymorphic among RBC genes [ 15 , 22 , 24 ], with growing GWAS evidence linking multiple variants to multiple traits [ 25 ]. Glycophorin loci are also thought to be ancient sites of positive and balancing selection, perhaps because of their roles in several infectious diseases [ 22 , 29 , 30 ].
Overall, it is plausible that the higher frequencies of protective alleles in ATP2B4, PIEZO1 , and GYPA/GYPB in sub-Saharan Africa, compared to the rest of the world, have been driven by selection consistent with the malaria hypothesis ( Box 1 ). Understanding the degree of protection that can be attributed to red cells, as opposed to other tissue types, will require more functional studies. Given the available evidence of pleiotropy ( Table 1 ), it is also possible that selective forces besides malaria have contributed to the observed frequency variation. Discovery and characterization of additional traits linked to these loci may aid in the design of novel treatments and preventative therapies for malaria.
Malaria susceptibility is highly polygenic
The largest GWAS of severe malaria to date sampled more than 17,000 individuals across 11 countries and 3 continents [ 7 ]. It successfully identified five genome-wide significant variants—in the genes HBB, GYPA/GYPB, ATP2B4, ABO , and an intergenic region on chromosome 6—that collectively explain ~11 % of the heritability of severe malaria risk. Aside from the identities of these specific variants, then, a major GWAS result is that the majority of heritability cannot yet be attributed to specific genomic loci ( Figure 1A, Key Figure ).
Figure 1, Key Figure. The main polygenic component of malaria resistance is not enriched in African populations.
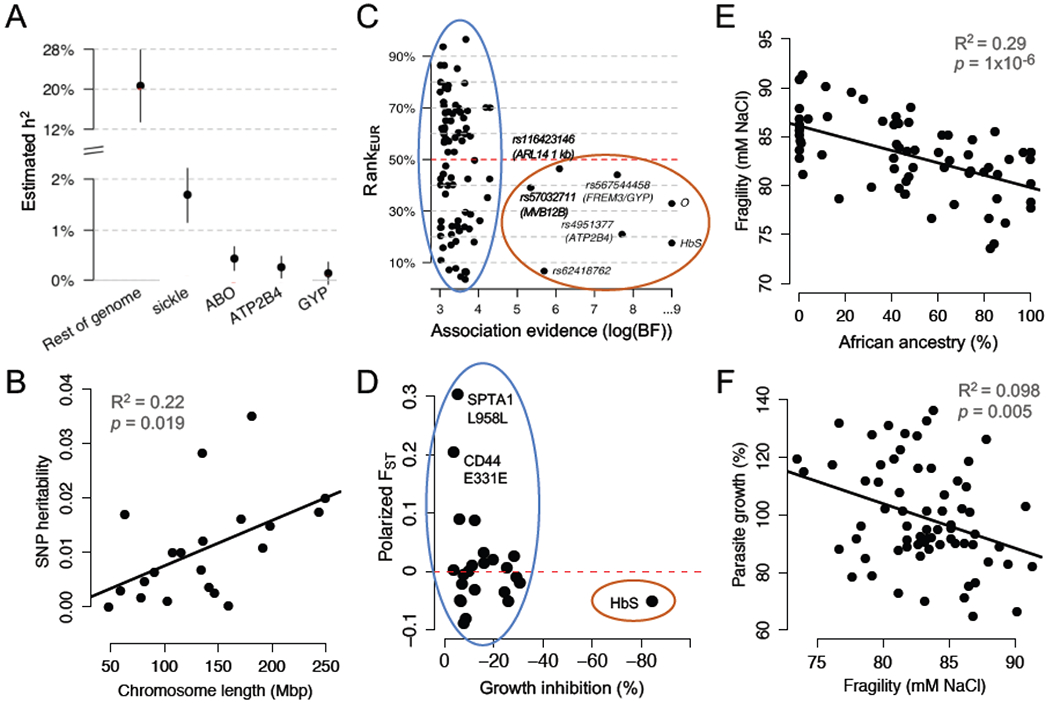
(A) 89% of the estimated heritability of severe malaria risk is attributed to variants with effects too small to be confidently detected by GWAS. Reproduced from [ 7 ]. (B) SNP-heritability of severe malaria risk is distributed across chromosomes proportional to their length. Replotted from [ 36 ]. (C) Strongly associated (large-effect) alleles from a GWAS for severe malaria risk occur at higher frequencies in African populations, compared to European populations (red oval). Smaller-effect alleles, which are less confidently associated, are equally likely to be more frequent in European populations (blue oval). Rank EUR is the conditional rank of the allele in European populations, given African allele count; values greater than/less than 50% indicate that the allele is more common in Europe/Africa. Reproduced from [ 7 ]. (D) Alleles from healthy RBCs associated with reduced P. falciparum growth rate in vitro are not enriched for higher frequencies in African populations (blue oval). Polarized F ST indicates the differentiation in allele frequencies between African and European populations in 1000 Genomes; values greater than/less than 0 indicate that the allele is more common in Europe/Africa. Replotted from [ 15 ]. (E) In donors who lack large-effect alleles for malaria resistance, red cell osmotic fragility is negatively related to exome-wide African ancestry. Replotted from [ 15 ]. (F) In donors who lack large-effect alleles for malaria resistance, higher red cell osmotic fragility predicts slower P. falciparum growth in vitro. Replotted from [ 15 ]. Panels A and C are reproduced with minor modifications under a Creative Commons license ( https://creativecommons.org/licenses/by/4.0/ ).
This “missing heritability” has a number of potential explanations that have been reviewed recently elsewhere [ 31 , 32 ]. As one example, GWAS have found variable associations between HbS and severe malaria because of variation in technical design (e.g. genotyping method) and environmental factors (e.g. local parasite genotype [ 33 ]). Furthermore, because Africans do not comprise a single, homogeneous population, detection of some variants (like Dantu) will depend on their local frequency and population structure. For these reasons, additional loci with large effects may remain to be identified by future studies, particularly those using African-centric SNP arrays or whole-genome sequencing.
Despite these limitations of GWAS, most authors still interpret large fractions of “missing heritability” as evidence that complex traits are highly polygenic [ 7 , 34 , 35 ]. From this perspective, nearly nine-tenths of the heritability of severe malaria risk could be attributed to variants with individual effects too small to be detected by GWAS of feasible size. Yet these variants appear to be so numerous that the sum of their tiny effects outweighs the much larger effects of the few most dramatic alleles ( Figure 1A ). A recent heritability analysis [ 36 ] demonstrated that the polygenic variation shaping severe malaria risk is distributed throughout the human genome, with each chromosome contributing a share of heritability roughly proportional to its physical size ( Figure 1B ). This diffuse genetic architecture resembles many other human traits, from red cell volume to type 1 diabetes [ 34 , 37 ]. How and whether polygenic variation may contribute to adaptation is an active area of interest in human population genetics [ 38 – 41 ]. Since severe malaria is well-established as a strong selective pressure ( Box 1 ), it offers a unique opportunity to contrast the evolution of protective alleles with small and large effects.
Polygenic variation linked to malaria protection is not enriched in African populations
The seven SNPs most strongly associated with malaria protection by GWAS ( Bayes factor (BF) > 100,000) are all more common in Africans than Europeans [ 7 ] ( Figure 1C , red oval). Although neither continental group represents a homogenous population, this pattern is consistent with the malaria hypothesis, which predicts local balancing selection of variants with large protective effects ( Box 1 ). In contrast, only 45 of the top 91 SNPs associated with malaria protection (BF > 1,000) are more common in Africans than Europeans [ 7 ] ( Figure 1C , blue oval). Although these weaker associations likely contain more false positives, this proportion is not meaningfully different from neutral (non-selective) expectations based on demography and population structure ( Box 2 ). Since SNPs with weaker evidence of association also tend to have smaller effect sizes , this result could suggest a distinct mode of evolution for the small-effect, polygenic variation underlying the majority of malaria susceptibility.
Our group recently found a similar result for RBC variants associated with P. falciparum growth in laboratory experiments [ 15 ]. Using RBCs sampled from healthy donors with a range of African ancestry, we identified 24 alleles (including PIEZO1 E756del and the ATP2B4 haplotype; Table 1 ) associated with reduced parasite growth. About half of these protective RBC variants are more frequent in Africans than Europeans ( Figure 1D , blue oval), which does not differ from expectations based on random exomic SNPs [ 15 ]. Notably, the sickle allele of hemoglobin (HbS) was the only single variant with a dramatic protective effect in these RBC assays ( Figure 1D , red oval). Although large-effect alleles from both GWAS and in vitro of studies do conform to the malaria hypothesis, it does not appear that small-effect alleles are, as a group, similarly differentiated between endemic and non-endemic populations.
Protective red cell traits are not enriched in African populations
Red blood cell phenotypes have been a constant focus of malaria research, from the initial formation of the malaria hypothesis ( Box 1 ) to modern GWAS of protective loci ( Table 1 ). Red cell traits related to volume, density, and hemoglobin content—known as red cell indices —are routinely measured in clinical settings around the globe. RBC membrane properties like fragility and deformability are also key to red cell function, blood storage, and diagnosis of hemolytic disease [ 5 , 8 ]. Most red cell traits have high heritability (40-90%), which large GWAS in multiple populations have attributed to polygenic variation [ 8 , 12 , 13 , 42 , 43 ].
Our recent study of healthy variation in RBCs without disease alleles (see Box 1 ) identified 10 polygenic traits, including RBC volume, that were strongly associated with P. falciparum growth in vitro [ 15 ]. Intriguingly, none of these protective traits (e.g. smaller RBC volume) were positively associated with African ancestry in non-carriers of disease alleles. Some RBC phenotypes were related to African ancestry, but not in the direction predicted by the malaria hypothesis. For example, African ancestry and African American ethnicity are strongly associated with decreased osmotic fragility of the RBC membrane [ 5 , 15 ] ( Figure 1E ). Membrane fragility, similar to “tension” measured by other studies [ 23 ], can be understood as an inverse of membrane deformability [ 15 , 44 ]. Our study found that lower osmotic fragility was associated with increased parasite growth [ 15 ] ( Figure 1F ), implying that RBCs with greater African ancestry are more susceptible to malaria parasites. Indeed, greater African ancestry in individuals without disease alleles predicted greater invasion efficiency of a clinical P. falciparum strain from Senegal [ 15 ].
An exaggerated protective effect of similar membrane properties has also been observed in African RBCs carrying the rare Dantu variant [ 23 ] ( Table 1 ). In that study, high-frame-rate video microscopy was used to simultaneously measure red cell tension and P. falciparum invasion, revealing that parasites preferentially invade RBCs with lower membrane tension. Accordingly, RBCs with higher baseline tension also experienced less membrane deformation when contacted by P. falciparum merozoites. Other recent studies have confirmed that RBC deformability upon merozoite contact is essential for, and correlated with, for P. falciparum invasion success [ 45 , 46 ]. Together, these data suggest that the low membrane fragility characteristic of RBCs with African ancestry makes them relatively more susceptible to P. falciparum parasites. It is possible that decreased membrane fragility serves another functional purpose, unrelated to malaria, that is yet to be discovered. We note that P. falciparum may adapt to the red cell traits that it usually encounters, more quickly than red cell traits themselves can evolve. Indeed, recent work has shown that that the protective effect of HbS varies by parasite genotype, consistent with HbS imposing selection on parasite alleles [ 33 ].
Lower red cell volume (MCV) has also been linked to reduced P. falciparum growth and/or invasion in diverse healthy adults [ 15 ], anemic African children [ 4 ], and chemically dehydrated RBCs [ 16 ]. Some earlier studies described lower MCV in African than European Americans, which has been attributed to selection for malaria resistance [ 4 ]. However, the deletion allele that causes alpha-thalassemia (see Box 1 ) strongly diminishes MCV [ 15 , 47 ] and is more common in African-Americans. When carriers of thalassemia are excluded, the apparent association between MCV and ancestry disappears, although the association between MCV and parasite growth remains [ 15 , 48 ]. While environmental variation can complicate inference of population-specific selection [ 49 ], Africans and Europeans might be expected to have distinct MCV optima, given differences in malaria prevalence and the protective value of low MCV. This lack of differentiation in a protective trait may suggest that other evolutionary forces are important for shaping polygenic variation in RBC volume, or that environmental factors have obscured the effects of selection.
One phenotype that has repeatedly and robustly been observed to differ between African- and European-Americans is hemoglobin concentration [ 50 ]. Polygenic, non-environmental factors appear to drive 6-7% lower blood hemoglobin concentration in African-Americans [ 50 ]. In malaria-endemic settings, this difference may be minor compared to anemia from non-genetic factors, which can reduce hemoglobin concentrations to 60% or less of reference values [ 51 ]. In 16 sub-Saharan African countries surveyed from 2010-2016, moderate to severe anemia was present in an average of 39.7% of children under 5 years of age [ 51 ]. Various recent studies have also found that hemoglobin concentration either improves [ 4 , 52 ], exacerbates [ 51 , 53 ], or has no effect [ 54 ] on malaria outcomes. This uncertainty, combined with the much larger impact of environment compared to genetics, raises the possibility that additional factors besides malaria pressure may have contributed to differences in hemoglobin concentration between Africans and Europeans.
Overall, there is currently little evidence that mean RBC trait differences between Africans and Europeans contribute to greater malaria resistance in Africans without known disease alleles. Since the RBC traits discussed here are strongly polygenic, this finding is consistent with the lack of differentiation of detectable, small-effect alleles between populations ( Figure 1C – D ). How can these conclusions be reconciled with evidence from large-effect alleles ( Box 1 ), which indicates strong selection to reduce P. falciparum growth in African RBCs?
Pervasive pleiotropy may limit polygenic adaptation to malaria
Many functional alleles, including those that confer malaria resistance ( Table 1 ; Box 1 ), have pleiotropic effects on multiple phenotypes. In recent years, the explosion of GWAS for human traits has enabled direct assessments of the extent of pleiotropy in the human genome. So far, 50-60% of SNPs that have been associated with one human trait are also associated with at least one more [ 55 , 56 ], consistent with widespread pleiotropy. A recent analysis of 558 GWAS traits found that overlapping, associated loci cover over half of the human genome, with over 90% of such loci associated with multiple traits [ 56 ]. In many cases, pleiotropic variants impact broadly expressed genes and signaling pathway components that are active in a multitude of cell types [ 55 ]. However, since SNPs identified by GWAS are usually in linkage disequilibrium with the true causal SNPs, it is theoretically possible that they tag multiple causal alleles with independent effects on multiple traits. We note that other limitations of GWAS, including technical explanations for missing heritability, also apply to these meta-analyses of multiple traits.
Four years ago, an influential model was proposed in which complex traits may be not just poly genic, but omni genic: affected by all genetic variants that impact gene expression in relevant cell types [ 34 ]. Such a model is plausible because biological networks, including transcriptional networks, are highly interconnected [ 57 ]. For example, a genetic variant that changes the transcription of one unit in a protein complex could consequently alter the folding, stability, physical interactions, or other activity of any number of functionally related proteins. The omnigenic model thus predicts extreme pleiotropy, even beyond what is directly observable from current GWAS data. It also helps explain why the heritability of most complex traits is spread so broadly throughout the genome ( Figure 1B ).
Understanding how abundant, functional variation is maintained within populations remains an ongoing challenge for evolutionary biologists [ 58 , 59 ]. If pleiotropy is so widespread that each functional allele has effects on many traits, then selection on all the traits—not just malaria resistance—could be expected to shape its allele frequency. We therefore propose that widespread pleiotropy may explain the current lack of observable differentiation between malaria-endemic and non-endemic populations for healthy variation that impacts malaria resistance ( Figure 1C – D ).
The red cell disorders that inspired the malaria hypothesis ( Box 1 ) are testaments to the strength of malaria selection, precisely because their large antimalarial effects outweigh their serious damage to other organismal phenotypes. These examples, however, may be exceptions. Most alleles associated with malaria protection have only small effects on individual risk ( Figure 1C – D ), but their combined effects far outweigh the handful of large-effect alleles in explaining malaria risk within a population ( Figure 1A ). Although malaria is a strong selective pressure, and RBC variation appears to impact malaria, we propose that the frequency of any functional allele will depend on its strength and direction of effect on many different traits—not only malaria resistance. Not all alleles that limit malaria should be expected to experience strong, population-specific selection; just as not every allele that is more common in Africa should be expected to impact malaria ( Box 2 ).
Concluding Remarks
Growing data from diverse human populations suggest that the malaria hypothesis may be a misleading heuristic for understanding global patterns of genetic variation, in part because malaria susceptibility is highly polygenic, and widespread pleiotropy may limit polygenic adaptation to malaria. This interpretation may be subject to change, however, as statistical and experimental methodologies improve. Current GWAS data are seriously limited by the large fraction of “missing heritability” for severe malaria risk ( Figure 1A ), but these limitations are expected to diminish as genetic diversity from non-European populations is discovered and incorporated into GWAS design. Similarly, steady increases in GWAS size; improved analysis of genetic correlations [ 38 , 40 , 41 ]; and steps toward controlling environmental variation [ 60 ] should enhance GWAS performance for small-effect, polygenic variation.
Our conclusions based on in vitro studies of P. falciparum and RBCs may also evolve as more physiologic models of this complex disease are developed, though this will require a substantial commitment of resources. For example, widespread adoption of improved humanized mouse models [ 61 ] will allow more cohesive and physiologic studies of severe malaria as a multi-organ disease. Increasingly detailed models of specific organ systems, such as the 3D architecture of the microvasculature [ 62 ] and blood-brain barrier [ 63 ], may also reveal non-RBC variation that is relevant to the malaria hypothesis. Still, the most direct test of the malaria hypothesis for polygenic variation may be large-scale, controlled infections of humans from diverse genetic backgrounds. At present, we have little knowledge of whether non-African populations contain their own large-effect variants for malaria resistance, nor whether the heritability of severe malaria varies around the world. A similar test could also be performed with genetically diverse non-human models, such as rodents sampled from wild populations.
As we await these developments, it is clear that continuing to explore aspects of malaria biology that do not necessarily adhere to the malaria hypothesis has the potential to advance our understanding of the pathogenesis of this complex disease. For example, RBC membrane dynamics clearly influence P. falciparum fitness [ 15 , 23 ], despite the higher frequency of susceptible phenotypes in African populations ( Figure 1E – F ). RBC membrane traits may represent a particularly exciting avenue for malaria prevention and therapeutics, given that RBC membrane variation is widespread and apparently well-tolerated in humans.
Casual invocation of the malaria hypothesis is not without costs, as its racial basis is known to reinforce misconceptions of biological heterogeneity within and across human groups [ 64 ] ( Box 2 ). Most genetic loci for malaria susceptibility likely have individually small effects, are spread across the genome, are pleiotropic for other traits, and are shared among many human populations. Despite evidence that protective alleles of large effect have experienced balancing selection, it is possible that polygenic variation shaping malaria susceptibility is constrained by correlation with other traits. In our opinion, a major goal of studying genetic susceptibility to disease is to identify novel biological pathways for therapeutic targeting (see Outstanding Questions ). Future research that embraces the polygenic, interconnected basis of complex traits will provide new insights into the progression of severe malaria.
Outstanding Questions.
What are the pleiotropic effects of the malaria-protective variants in ATP2B4, PIEZO1 , and GYPA/GYPB?
Besides red blood cells, which tissues express genes that contribute to the heritability of severe malaria?
What traits are genetically correlated with severe malaria risk?
How many common variants adhering to the malaria hypothesis remain to be identified, particularly as knowledge of African genetic diversity improves?
Would larger GWAS or in vitro studies yield more robust signals of polygenic adaptation to malaria?
Why are red cell traits so variable within and across human populations?
Is common red cell variation important for P. falciparum parasites in vivo ? How does common red cell variation contribute to severe malaria?
Highlights.
In malaria-endemic populations, the malaria hypothesis predicts balancing selection on protective loci that interfere with red blood cell (RBC) function.
Novel variants in the RBC genes PIEZO1, ATP2B4 , and GYPA/GYPB are associated with strong protection from severe malaria and are more common in Africans than Europeans, consistent with the malaria hypothesis.
GWAS attribute most of the heritability of severe malaria risk to many loci with small effects spread across the genome.
African populations are not enriched for small-effect alleles associated with protection from severe malaria. Similarly, healthy RBC traits associated with reduced P. falciparum growth in vitro are not positively associated with African ancestry.
Pervasive pleiotropy may limit adaptation in complex traits. Therefore, host variation that impacts malaria may not necessarily conform to the malaria hypothesis.
Acknowledgements
We thank Abigail LaBella, Grant Kinsler, Sophie Walton, and members of the Egan lab for helpful discussion and comments. E.R.E. is supported by an NSF Postdoctoral Research Fellowship in Biology (Grant No. 2109851). This work was supported by NIH grant 1DP2HL13718601 to E.S.E., who is a Tashia and John Morgridge Endowed Faculty Scholar in Pediatric Translational Medicine through the Stanford Maternal Child Health Research Institute.
a condition in which red blood cell quantity or hemoglobin concentration is lower than normal.
the active maintenance of multiple alleles at one genetic locus.
a ratio of the likelihood of one hypothesis over another. In GWAS, larger values are interpreted as stronger evidence of association between an allele and a trait, compared to a null hypothesis of no association.
a sharp decline in population size that reduces genetic diversity.
a phenotype influenced by many genetic and environmental factors.
a change in allele frequency due to chance, such as random sampling of chromosomes during meiosis. Drift has stronger impacts in smaller populations.
when estimated from an association study, the coefficient of an allele in a linear regression model of a trait.
a relative, average measure of the genetic contribution to the next generation from individuals with a certain genotype or phenotype.
the production of sexual cells. In P. falciparum , gametocytes produced from asexual blood stages are essential to mosquito transmission.
an association between a trait and allele with p <5x10 −8 , based on a Bonferroni correction of all common independent SNPs in the human genome. Alleles with larger effect sizes and higher frequency are more likely to be genome-wide significant.
an observational study of the association between a trait and genome-wide genetic markers, usually conducted in many individuals.
the fraction of variance in a trait explained by additive genetic variation, as opposed to environment and measurement error.
correlation between alleles within a population, usually on the same physical chromosome.
not selected; having no impact on fitness.
the likelihood of an outcome for one group relative to another. For example, the likelihood of severe malaria for individuals with a particular allele.
influenced by all genes expressed in cell types relevant to a trait. The omnigenic model proposes that expression of a trait’s ‘core’ genes is regulated by many ‘peripheral’ genes in an interconnected network, allowing any variant within that network to have indirect effects on the trait.
the propensity of a red cell to lyse while absorbing water under hypotonic stress, which reflects biophysical membrane properties.
traditionally, influenced by more than one gene. Today, this term is commonly used for traits influenced by hundreds or thousands of genetic variants, each with a small effect on total trait variation.
the organization of genetic variation among populations, driven by the evolutionary processes of mutation, recombination, drift, and selection.
(of an allele) influencing two or more traits.
standardized blood tests of red cell count, size, hemoglobin content, and variability.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
- 1. Shriner D and Rotimi CN (2018) Whole-Genome-Sequence-Based Haplotypes Reveal Single Origin of the Sickle Allele during the Holocene Wet Phase. Am. J. Hum. Genet 102, 547–556 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 2. World Health Organization (2020) World malaria report 2020: 20 years of global progress and challenges, World Health Organization [ Google Scholar ]
- 3. Kariuki SN and Williams TN (2020) Human genetics and malaria resistance. Hum. Genet 139, 801–811 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 4. Goheen MM et al. (2016) Anemia Offers Stronger Protection Than Sickle Cell Trait Against the Erythrocytic Stage of Falciparum Malaria and This Protection Is Reversed by Iron Supplementation. EBioMedicine 14, 123–130 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 5. Kanias T et al. (2017) Ethnicity, sex, and age are determinants of red blood cell storage and stress hemolysis: Results of the REDS-III RBC-Omics study. Blood Adv. 1, 1132–1141 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 6. Ma S et al. (2018) Common PIEZO1 Allele in African Populations Causes RBC Dehydration and Attenuates Plasmodium Infection. Cell 173, 443–455.e12 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 7. Network MGE (2019) Insights into malaria susceptibility using genome-wide data on 17,000 individuals from Africa, Asia and Oceania. Nat. Commun 10, 5732. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 8. Page GP et at. (2021) Multiple-ancestry genome-wide association study identifies 27 loci associated with measures of hemolysis following blood storage. J. Clin. Invest 131, [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 9. Zhang DL et al. (2018) Erythrocytic ferroportin reduces intracellular iron accumulation, hemolysis, and malaria risk. Science 359, 1520–1523 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 10. Biddanda A et al. (2020) A variant-centric perspective on geographic patterns of human allele frequency variation. eLife 9, e60107. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 11. Zámbó B et al. (2017) Decreased calcium pump expression in human erythrocytes is connected to a minor haplotype in the ATP2B4 gene. Cell Calcium 65, 73–79 [ DOI ] [ PubMed ] [ Google Scholar ]
- 12. Hodonsky CJ et al. (2018) Generalization and fine mapping of red blood cell trait genetic associations to multi-ethnic populations: The PAGE study. Am. J. Hematol 93, 1061–1073 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 13. Astle WJ et al. (2016) The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell 167, 1415–1429.e19 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 14. Lessard S et al. (2017) An erythroid-specific ATP2B4 enhancer mediates red blood cell hydration and malaria susceptibility. J. Clin. Invest 127, 3065–3074 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 15. Ebel ER et al. (2021) Common host variation drives malaria parasite fitness in healthy human red cells. eLife 10, e69808. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 16. Tiffert T et al. (2005) The hydration state of human red blood cells and their susceptibility to invasion by Plasmodium falciparum. Blood 105, 4853–4860 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 17. Nguetse CN et al. (2020) A common polymorphism in the mechanosensitive ion channel PIEZO1 is associated with protection from severe malaria in humans. Proc. Natl. Acad. Sci 117, 9074–9081 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 18. Ilboudo Y et al. (2018) A common functional PIEZO1 deletion allele associates with red blood cell density in sickle cell disease patients. Am. J. Hematol 93, E362–E365 [ DOI ] [ PubMed ] [ Google Scholar ]
- 19. Rooks H et al. (2019) A gain of function variant in PIEZO1 (E756del) and sickle cell disease. Haematologica 104, e91–e93 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 20. Thye T et al. (2021) Human genetic variant E756del in the ion channel PIEZO1 not associated with protection from severe malaria in a large Ghanaian study. J. Hum. Genet DOI: 10.1038/s10038-021-00958-2 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 21. Jaskiewicz E et al. (2019) Erythrocyte glycophorins as receptors for Plasmodium merozoites. Parasit. Vectors 12, 317. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 22. Leffler EM et al. (2017) Resistance to malaria through structural variation of red blood cell invasion receptors. Science 356, [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 23. Kariuki SN et al. (2020) Red blood cell tension protects against severe malaria in the Dantu blood group. Nature 585, 579–583 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 24. Algady W et al. (2018) The Malaria-Protective Human Glycophorin Structural Variant DUP4 Shows Somatic Mosaicism and Association with Hemoglobin Levels. Am. J. Hum. Genet 103, 769–776 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 25. Buniello A et al. (2019) The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 26. Campino S et al. (2016) Genomic variation in two gametocyte non-producing Plasmodium falciparum clonal lines. Malar. J 15, 229. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 27. Passini FS et al. (2021) Shear-stress sensing by PIEZO1 regulates tendon stiffness in rodents and influences jumping performance in humans. Nat. Biomed. Eng DOI: 10.1038/s41551-021-00716-x [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 28. Ma S et al. (2021) A role of PIEZO1 in iron metabolism in mice and humans. Cell 184, 969–982.e13 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 29. Bigham AW et al. (2018) Complex signatures of natural selection at GYPA. Hum. Genet 137, 151–160 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 30. Johnson KE and Voight BF (2018) Patterns of shared signatures of recent positive selection across human populations. Nat. Ecol. Evol 2, 713–720 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 31. Génin E (2020) Missing heritability of complex diseases: case solved? Hum. Genet 139, 103–113 [ DOI ] [ PubMed ] [ Google Scholar ]
- 32. Young AI (2019) Solving the missing heritability problem. PLOS Genet. 15, e1008222. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 33. Band G et al. (2021) Malaria protection due to sickle haemoglobin depends on parasite genotype. Nature DOI: 10.1038/s41586-021-04288-3 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 34. Boyle EA et al. (2017) An expanded view of complex traits: from polygenic to omnigenic. Cell 169, 1177–1186 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 35. Crouch DJM and Bodmer WF (2020) Polygenic inheritance, GWAS, polygenic risk scores, and the search for functional variants. Proc. Natl. Acad. Sci 117, 18924–18933 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 36. Damena D and Chimusa ER (2020) Genome-wide heritability analysis of severe malaria resistance reveals evidence of polygenic inheritance. Hum. Mol. Genet 29, 168–176 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 37. Shi H et al. (2016) Contrasting the Genetic Architecture of 30 Complex Traits from Summary Association Data. Am. J. Hum. Genet 99, 139–153 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 38. Berg JJ et al. (2019) Reduced signal for polygenic adaptation of height in UK Biobank. eLife 8, e39725. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 39. Racimo F et al. (2018) Detecting Polygenic Adaptation in Admixture Graphs. Genetics 208, 1565–1584 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 40. Sohail M et al. (2019) Polygenic adaptation on height is overestimated due to uncorrected stratification in genome-wide association studies. eLife 8, e39702. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 41. Stern AJ et al. (2021) Disentangling selection on genetically correlated polygenic traits via whole-genome genealogies. Am. J. Hum. Genet 108, 219–239 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 42. Chen M-FI. et al. (2020) Trans-ethnic and Ancestry-Specific Blood-Cell Genetics in 746,667 Individuals from 5 Global Populations. Cell 182, 1198–1213.e14 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 43. Vuckovic D et al. (2020) The Polygenic and Monogenic Basis of Blood Traits and Diseases. Cell 182, 1214–1231.e11 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 44. Matthews K et al. (2017) Microfluidic analysis of red blood cell deformability as a means to assess hemin-induced oxidative stress resulting from Plasmodium falciparum intraerythrocytic parasitism. Integr. Biol 9, 519–528 [ DOI ] [ PubMed ] [ Google Scholar ]
- 45. Blake TCA et al. (2020) Actomyosin forces and the energetics of red blood cell invasion by the malaria parasite Plasmodium falciparum. PLOS Pathog. 16, e1009007. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 46. Sisquella X et al. (2017) Plasmodium falciparum ligand binding to erythrocytes induce alterations in deformability essential for invasion. eLife 6, e21083. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 47. Nunchai C et al. (2020) Optimal cutoff of mean corpuscular volume (MCV) for screening of alpha-thalassemia 1 trait. J. Obstet. Gynaecol. Res 46, 774–778 [ DOI ] [ PubMed ] [ Google Scholar ]
- 48. Beutler E and West C (2005) Hematologic differences between African-Americans and whites: the roles of iron deficiency and alpha-thalassemia on hemoglobin levels and mean corpuscular volume. Blood 106, 740–745 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 49. Harpak A and Przeworski M (2021) The evolution of group differences in changing environments. PLOS Biol. 19, e3001072. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 50. Kang W et al. (2021) Ethnic Differences in Iron Status. Adv. Nutr DOI: 10.1093/advances/nmab035 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 51. Papaioannou I et al. (2019) Malaria-anemia comorbidity prevalence as a measure of malaria-related deaths in sub-Saharan Africa. Sci. Rep 9, 11323. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 52. Goheen MM et al. (2017) Host iron status and erythropoietic response to iron supplementation determines susceptibility to the RBC stage of falciparum malaria during pregnancy. Sci. Rep 7, 17674. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 53. Lombardo P et al. (2017) Hemoglobin Levels and the Risk of Malaria in Papua New Guinean Infants: A Nested Cohort Study. Am. J. Trop. Med. Hyg 97, 1770–1776 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 54. Muriuki JM et al. (2019) Iron Status and Associated Malaria Risk Among African Children. Clin. Infect. Dis 68, 1807–1814 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 55. Shikov AE et al. (2020) Phenome-wide functional dissection of pleiotropic effects highlights key molecular pathways for human complex traits. Sci. Rep 10, 1037. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 56. Watanabe K et al. (2019) A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet 51, 1339–1348 [ DOI ] [ PubMed ] [ Google Scholar ]
- 57. Singh RS and Gupta BP (2020) Genes and genomes and unnecessary complexity in precision medicine. Npj Genomic Med. 5, 1–9 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 58. Llaurens V et al. (2017) Genetic architecture and balancing selection: the life and death of differentiated variants. Mol. Ecol 26, 2430–2448 [ DOI ] [ PubMed ] [ Google Scholar ]
- 59. Bertram J and Masel J (2019) Different mechanisms drive the maintenance of polymorphism at loci subject to strong versus weak fluctuating selection. Evolution 73, 883–896 [ DOI ] [ PubMed ] [ Google Scholar ]
- 60. Watson JA et al. (2021) Improving statistical power in severe malaria genetic association studies by augmenting phenotypic precision. eLife 10, e69698. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 61. Minkah NK et al. (2018) Humanized Mouse Models for the Study of Human Malaria Parasite Biology, Pathogenesis, and Immunity. Front. Immunol 9, 807. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 62. Bernabeu M et al. (2021) Bioengineered 3D Microvessels for Investigating Plasmodium falciparum Pathogenesis. Trends Parasitol. 37, 401–413 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 63. Long RKM et al. (2021) Understanding parasite–brain microvascular interactions with engineered 3D blood–brain barrier models. Mol. Microbiol 10.1111/mmi.14852 [ DOI ] [ PubMed ] [ Google Scholar ]
- 64. Sparks RA et al. (2020) Using Culturally Relevant Pedagogy to Reconsider the Genetics Canon. J. Microbiol. Biol. Educ 21, [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 65. Haldane JBS (1949) THE RATE OF MUTATION OF HUMAN GENES. Hereditas 35, 267–273 [ Google Scholar ]
- 66. Allison AC (1954) The distribution of the sickle-cell trait in East Africa and elsewhere, and its apparent relationship to the incidence of subtertian malaria. Trans. R. Soc. Trop. Med. Hyg 48, 312–318 [ DOI ] [ PubMed ] [ Google Scholar ]
- 67. Allison AC (1954) Protection Afforded by Sickle-cell Trait Against Subtertian Malarial Infection. BMJ 1, 290–294 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 68. Jamrozik E and Selgelid MJ (2021) History of Human Challenge Studies. In Human Challenge Studies In Endemic Settings: Ethical and Regulatory Issues (Jamrozik E and Selgelid MJ, eds), pp. 9–23, Springer International Publishing [ Google Scholar ]
- 69. Archer NM et al. (2018) Resistance to Plasmodium falciparum in sickle cell trait erythrocytes is driven by oxygen-dependent growth inhibition. Proc. Natl. Acad. Sci 115, 7350–7355 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 70. Ha J et al. (2019) Hemoglobin E, malaria and natural selection. Evol. Med. Public Health 2019, 232–241 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 71. Clarke GM et al. (2017) Characterisation of the opposing effects of G6PD deficiency on cerebral malaria and severe malarial anaemia. eLife 6, e15085. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 72. Paquette AM et al. (2015) The evolutionary origins of Southeast Asian Ovalocytosis. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis 34, 153–159 [ DOI ] [ PubMed ] [ Google Scholar ]
- 73. Tétard M et al. (2017) Heterozygous HbAC but not HbAS is associated with higher newborn birthweight among women with pregnancy-associated malaria. Sci. Rep 7, 1414. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 74. Bergström A et al. (2021) Origins of modern human ancestry. Nature 590, 229–237 [ DOI ] [ PubMed ] [ Google Scholar ]
- 75. Rosenberg NA (2011) A Population-Genetic Perspective on the Similarities and Differences among Worldwide Human Populations. Hum. Biol 83, 659–684 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 76. Coop G et al. (2009) The Role of Geography in Human Adaptation. PLoS Genet. 5, e1000500. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 77. Lewontin RC (1972) The Apportionment of Human Diversity. In Evolutionary Biology: Volume 6 (Dobzhansky T et al. , eds), pp. 381–398, Springer US [ Google Scholar ]
- 78. Rosenberg NA et al. (2019) Interpreting polygenic scores, polygenic adaptation, and human phenotypic differences. Evol. Med. Public Health 2019, 26–34 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 79. Karczewski KJ et al. (2020) The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 80. Ndila CM et al. (2018) Human candidate gene polymorphisms and risk of severe malaria in children in Kilifi, Kenya: a case-control association study. Lancet Haematol. 5, e333–e345 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- View on publisher site
- PDF (329.6 KB)
- Collections
Similar articles
Cited by other articles, links to ncbi databases.
- Download .nbib .nbib
- Format: AMA APA MLA NLM
Add to Collections
An official website of the United States government
Official websites use .gov A .gov website belongs to an official government organization in the United States.
Secure .gov websites use HTTPS A lock ( Lock Locked padlock icon ) or https:// means you've safely connected to the .gov website. Share sensitive information only on official, secure websites.
- Publications
- Account settings
- Advanced Search
- Journal List
How Malaria Has Affected the Human Genome and What Human Genetics Can Teach Us about Malaria
Dominic p kwiatkowski.
- Author information
- Article notes
- Copyright and License information
Address for correspondence and reprints: Dr. Dominic P. Kwiatkowski, Wellcome Trust Centre for Human Genetics, Oxford OX3 7BN, United Kingdom. E-mail: [email protected]
Received 2005 Jun 3; Accepted 2005 Jun 3; Issue date 2005 Aug.
Malaria is a major killer of children worldwide and the strongest known force for evolutionary selection in the recent history of the human genome. The past decade has seen growing evidence of ethnic differences in susceptibility to malaria and of the diverse genetic adaptations to malaria that have arisen in different populations: epidemiological confirmation of the hypotheses that G6PD deficiency, α + thalassemia, and hemoglobin C protect against malaria mortality; the application of novel haplotype-based techniques demonstrating that malaria-protective genes have been subject to recent positive selection; the first genetic linkage maps of resistance to malaria in experimental murine models; and a growing number of reported associations with resistance and susceptibility to human malaria, particularly in genes involved in immunity, inflammation, and cell adhesion. The challenge for the next decade is to build the global epidemiological infrastructure required for statistically robust genomewide association analysis, as a way of discovering novel mechanisms of protective immunity that can be used in the development of an effective malaria vaccine.
Introduction
One of the most important causes of child mortality worldwide is the malaria parasite Plasmodium falciparum, which annually kills >1 million children in Africa alone. This death toll is only one aspect of the global burden of malaria. P. falciparum is estimated to cause about half a billion episodes of disease each year (Snow et al. 2005 ), and there are hundreds of millions of cases due to other parasite species— Plasmodium vivax, Plasmodium malariae, and Plasmodium ovale. In regions of high malaria transmission, every member of the community might be chronically infected (Trape et al. 1994 ), and, in one Gambian village, it was found that about one-quarter of all children with severe complications of the disease were admitted to the hospital during the first 10 years of life (Snow et al. 1997 ).
When malaria's effect on child mortality is considered—and it was probably even greater before antimalarial drugs and other control measures were introduced—it is not surprising that malaria is the strongest known selective pressure in the recent history of the human genome. Malaria is the evolutionary driving force behind sickle-cell disease, thalassemia, glucose-6-phosphatase deficiency, and other erythrocyte defects that together comprise the most common Mendelian diseases of humankind. HbS, the allele that gives rise to sickle hemoglobin, is regarded as the classic paradigm of balanced polymorphisms in human populations (Feng et al. 2004 ; Hedrick 2004 ). It is a variant of the HBB gene (which encodes β-globin) that has arisen independently in different locations and is maintained at ∼10% frequency in many malaria-endemic regions (Flint et al. 1998 ). HbS homozygotes suffer sickle-cell disease, but heterozygotes have a 10-fold reduced risk of severe malaria (Hill et al. 1991 ; Ackerman et al. 2005 ).
What is remarkable is the range of erythrocyte variants, apart from HbS, that have resulted from evolutionary selection by malaria. They include other variants of the HBB gene—namely, HbC and HbE (Agarwal et al. 2000 ; Modiano et al. 2001 b ; Chotivanich et al. 2002 ; Ohashi et al. 2004 ); regulatory defects of HBA and HBB, which cause α and β thalassemia (Flint et al. 1986 ; Williams et al. 1996 ; Allen et al. 1997 ), variation in the structural protein SLC4A1, which causes ovalocytosis (Foo et al. 1992 ; Genton et al. 1995 ; Allen et al. 1999 ); variation in the chemokine receptor FY, which causes the Duffy-negative blood group (Miller et al. 1976 ; Chitnis and Miller 1994 ; Tournamille et al. 1995 ; Hamblin and Di Rienzo 2000 ); and polymorphisms of the red-cell enzyme gene G6PD, which causes glucose-6-phosphate dehydrogenase deficiency (Bienzle et al. 1972 ; Ganczakowski et al. 1995 ; Ruwende and Hill 1998 ; Tishkoff et al. 2001 ; Sabeti et al. 2002 b ).
This is probably only the tip of the iceberg. Surprisingly little is currently known about the effects of malaria on the evolution of the human immune system, possibly because the phenotypic consequences are more subtle than those of the classic erythrocyte variants; for example, alteration of a splenic dendritic cell receptor is not as easy to visualize as a sickling red cell. However, the last few years have seen a rapid growth in the number of reported genetic associations with susceptibility and resistance to malaria, many of which involve immune system and inflammatory genes.
The purpose of this review is to provide an overview of what is currently known about genetic resistance to malaria and to highlight directions that are likely to see major advances in the next few years.
Evolutionary Selection by Malaria
Evolutionary selection by malaria (reviewed by Flint et al. [ 1998 ] and Tishkoff and Williams [ 2002 ]) is remarkable in two respects. First, the selective pressure is very strong; this is evident from the fact that the HbS allele has risen to high frequencies in malaria-exposed populations despite the fatal consequences for homozygotes. Second, different populations have developed independent evolutionary responses to malaria, and this is seen at both the global and the local levels. The most striking example is the HBB gene, in which three different coding SNPs confer protection against malaria: Glu6Val (HbS), Glu6Lys (HbC), and Glu26Lys (HbE). The HbS allele is common in Africa but rare in Southeast Asia, whereas the opposite is true for the HbE allele. However, a more complex picture emerges at the local level, exemplified by the Dogon people of Mali, who have a much lower frequency of the HbS allele than do most other West African groups and instead have a high frequency of the HbC allele (Agarwal et al. 2000 ). A further level of complexity is that, within Africa, the HbS allele is found in four distinct haplotypes (Chebloune et al. 1988 ; Nagel and Ranney 1990 ; Lapoumeroulie et al. 1992 ), a finding that has been generally interpreted to imply that the same mutation has arisen independently in four different Africa populations, although it has been pointed out that there may be other explanations (Flint et al. 1998 ). The different geographic distributions of α thalassemia, G6PD deficiency, ovalocytosis, and the Duffy-negative blood group are further examples of the general principle that different populations have evolved different genetic variants to protect against malaria.
The fact that different malaria-resistance alleles have arisen in different places suggests that a great deal of evolutionary selection by malaria has happened relatively recently in human history and certainly since humans started to migrate out of Africa. This is supported by analyses of recent positive selection in the human genome. Haplotype analysis and statistical modeling of an African malaria-resistance allele at the G6PD locus suggests an origin within the last 10,000 years or so (Tishkoff et al. 2001 ), whereas analysis of the Southeast Asian HbE allele suggests that it originated within the last 5,000 years (Ohashi et al. 2004 ). Studies of G6PD and CD40L malaria-resistance alleles in West Africa that made use of the long-range haplotype test are also consistent with recent positive selection (Sabeti et al. 2002 b ).
Another line of evidence comes from population-genetics analysis of malaria parasites. Malaria parasites existed long before humans—there are different Plasmodium species that infect birds, lizards, rodents, and other primates—but what geneticists would like to know is the timepoint at which the unusually virulent species P. falciparum began to expand in human populations. This has been addressed by a variety of approaches, with conflicting results (reviewed by Conway [ 2003 ] and Hartl [ 2004 ]), but the most persuasive evidence comes from a detailed analysis of 100 mtDNA sequences sampled from around the world (Joy et al. 2003 ). This suggests that some forms of P. falciparum may have existed 100,000 years ago, but that the African malaria parasite population suddenly increased ∼10,000 years ago and subsequently spread to other regions. This observation, together with analysis of the speciation of human malaria vectors by polytene chromosome analysis (Coluzzi 1999 ; Coluzzi et al. 2002 ), is consistent with the hypothesis that the emergence of P. falciparum as a major human pathogen coincides with the beginnings of agriculture, when human populations started to form resident communities that allowed the establishment of a substantial reservoir of infection.
These findings are not just of historical interest, they may also be of practical value in the search for novel malaria-resistance loci. One of the major challenges confronting genomewide association analysis is determining how to select the most-efficient set of markers to analyze. In the case of malaria, we are particularly interested in alleles that show evidence of recent positive selection in regions where malaria is endemic. P. falciparum seems to have emerged as a powerful selective force subsequent to the divergence of African, Asian, and European populations, so an obvious starting point for genomewide analysis of malaria-resistance loci is to screen alleles that show large differences in frequency between major population groups.
Complex Genetic Basis of Resistance to Malaria
The genetic basis of resistance to malaria is complex at several levels. It is likely that many different genes are involved and that they interact with environmental variables and with parasite genetic factors. Here, we consider some further complexities of studying genetic resistance to malaria—namely, the range of phenotypes involved, the practical difficulties of studying families, and the remarkable geographic and ethnic heterogeneity of malaria-resistance factors.
Susceptibility to and resistance to malaria can be measured in several ways. Usually, they are studied in regions with high levels of malaria transmission. When everyone is repeatedly bitten by infected mosquitoes, many children and adults are likely to have parasitemia (parasites in the blood), children may have two or three episodes of malaria fever each year, and a small minority (e.g., 1%) of malaria-fever episodes lead to severe malaria (i.e., death or life-threatening complications due to malaria). Different genetic factors may determine the risk of an exposed person for developing parasitemia, the risk of a parasitemic person for becoming ill with malaria fever, and the risk of a person with malaria fever for developing severe malaria. Parasitemia and fever can be regarded as quantitative phenotypes that are ascertained by repeated measurements within the community, whereas severe malaria is a qualitative phenotype that is typically ascertained in hospital-based studies. In principle, studies of severe malaria would be expected to detect genetic factors at each stage in the causal chain of disease progression.
Severe malaria is the phenotype that matters most to vaccine developers and to those interested in evolutionary selection. Severe malaria is composed of a number of subphenotypes that may occur alone or in combination. In African children, these are cerebral malaria, severe malarial anemia, and respiratory distress (Marsh et al. 1995 ). When we recruit 100 children with severe malaria who are in the hospital to a study, we are effectively identifying those, of perhaps 10,000 children who live in the hospital’s catchment area, with the lowest levels of protection, and this tends to enrich the sample for strong genetic effects. For example, HbS heterozygotes have an ∼2-fold reduction in the risk of malaria-fever episodes but a 10-fold reduction in the risk of severe malaria (Hill et al. 1991 ).
Family Studies
It is difficult to get an accurate measure of the familial component of resistance to severe malaria. Cases of severe malaria are relatively easy to identify in hospitals, but the transient nature of the illness makes it difficult to ascertain the affected status of relatives, particularly in communities that lack detailed medical records and in which there are other infectious causes of child mortality that can be confused with malaria. By careful questioning of families of affected individuals, a recent study in Mali estimated a sibling risk (λ s ) of 2.5 for cerebral malaria and 4.9 for severe malarial anemia (Ranque et al. 2005 ). This, of course, does not distinguish genetic effects from those of shared environment.
To do an accurate family study of malaria, it is necessary to perform detailed longitudinal analysis, which, in practice, means studying a relatively small number of individuals, such as a set of twins or a village of a few hundred people. Such studies lack power to investigate severe malaria, which is a relatively rare event, but they can evaluate quantitative traits such as the level of parasitemia or frequency of malaria-fever episodes. The fact that the level of parasitemia and the frequency of malaria fever both decline markedly with age must be factored into the analysis. A longitudinal study of Gambian twins showed that susceptibility to malaria-fever episodes is determined partly by genetic factors (Jepson et al. 1995 ), with linkage to the major histocompatibility complex (MHC) region on chromosome 6 (Jepson et al. 1997 ). A series of longitudinal family studies of parasitemia have been performed in Cameroon and Burkina Faso. An initial segregation analysis suggested the involvement of a major gene that controls blood infection levels (Abel et al. 1992 ), and this hypothesis seemed to be supported by a somewhat bimodal distribution of parasite density levels among pregnant women, who are particularly susceptible to malaria (Cot et al. 1993 ). However, subsequent studies indicate that complex genetic factors are involved (Garcia et al. 1998 a ; Rihet et al. 1998 a ) and that there is linkage to the MHC and the 5q31-33 region (Garcia et al. 1998 b ; Rihet et al. 1998 b ; Flori et al. 2003 ). A longitudinal family analysis in Sri Lanka concluded that there were consistent individual differences in susceptibility to clinical malaria episodes, of which about one-half appeared to have a genetic basis (Mackinnon et al. 2000 ).
It is also possible to estimate the genetic component of individual variation in immunological responses. A twin study in Liberia found evidence of heritability in antimalarial antibody responses that did not appear to be determined by HLA class II genes (Sjoberg et al. 1992 ). Familial segregation analysis of immunological responses to malaria antigens in Papua New Guinea has suggested that Mendelian effects might govern specific antigen responses, but the overall picture is complex (Stirnadel et al. 1999 a , 2000 a , 2000 b ).
Ethnic Differences
One of the most striking examples of differential disease susceptibility among human populations is the complete resistance of most of the population of sub-Saharan Africa to P. vivax infection, whereas all other human populations are vulnerable to this species of malaria parasite. This resistance is due to a SNP in the FY gene that results in the Duffy blood group–negative phenotype (Miller et al. 1976 ; Tournamille et al. 1995 ); the precise protective mechanism is discussed below (see the “ Malaria and the Red Cell ” section). Thus, whereas P. vivax infection is common in Asia and South America and used to be widely distributed throughout Europe, it is extremely rare in most of sub-Saharan Africa, although all the right environmental conditions exist for it to be transmitted there.
Striking differences in resistance to malaria have also been observed among ethnic groups who live in the same area. The Tharu people, who inhabit the malarious Terai region of Nepal, have a much lower prevalence of malaria than do other ethnic groups in the same region (Terrenato et al. 1988 ), and this may possibly be explained by the extremely high frequency of α thalassemia in the Tharu population (Modiano et al. 1991 ).
The Fulani people are traditionally nomadic pastoral people who are found across West Africa, often settled in close proximity to other ethnic groups. Studies in Burkina Faso (Modiano et al. 1996 ) and, more recently, in Mali (Dolo et al. 2005 ) have documented a significantly lower prevalence of malaria parasitemia and fewer clinical attacks of malaria among the Fulani than among other ethnic groups who live in neighboring villages. The Fulani have a distinctive culture, but detailed epidemiological investigations indicate that their resistance to malaria arises primarily from genetic factors. Importantly, it has also been observed that the Fulani have high levels of antimalarial antibodies (Modiano et al. 1998 , 1999 ) and a low frequency of protective globin variants and other classic malaria-resistance factors (Modiano et al. 2001 a ). There is therefore much interest in discovering the genetic factors that determine the high antibody responses seen in the Fulani; the possible role of the IL4 gene is discussed below (see the “ Antibody Response ” section) (Luoni et al. 2001 ; Farouk et al. 2005 ).
Malaria and the Red Cell
Erythrocyte surface.
Many important things happen at the erythrocyte surface in malaria ( table 1 ). The parasite binds to erythrocyte surface molecules as the first stage in a complex and marvelous series of events—still poorly understood—that gets the parasite into the erythrocyte without destroying it (Sibley 2004 ). Once inside the erythrocyte, the parasite manufactures a set of proteins that it sends to the cell surface (Kyes et al. 2001 ). Some of these parasite-derived erythrocyte-membrane proteins bind to endothelial adhesion molecules and thereby cause parasitized erythrocytes to sequester in small blood vessels; this is thought to be a strategy for immune evasion, since it prevents the parasites from having to circulate through the spleen. But these parasite-derived molecules on the erythrocyte surface are themselves targets for immunological attack, which they counter with an extraordinary capacity for antigenic variation. We discuss in the “ Cytoadherence, a Major Factor in Malaria Pathogenesis ” section how human genetic variation influences endothelial cytoadherence; here, we focus on how it affects erythrocyte invasion.
Common Erythrocyte Variants That Affect Resistance to Malaria
The study of human genetics uncovered a key step in the molecular process of erythrocyte invasion by P. vivax. The Duffy antigen, encoded by the FY gene, is a chemokine receptor that is expressed in various cell types. The Duffy antigen is expressed in erythrocytes in most populations (but not in sub-Saharan Africa) that have a promoter SNP that disrupts a binding site for the erythroid transcription factor GATA-1 (Tournamille et al. 1995 ). This completely prevents P. vivax from invading erythrocytes, and it accounts for the remarkable absence of P. vivax in parts of Africa in which other species of malaria parasite are extremely common (Miller et al. 1976 ). It was this genetic discovery that led to the discovery of the P. vivax Duffy-binding protein, a parasite molecule that is critical for erythrocyte invasion by P. vivax (Chitnis and Miller 1994 ) and is now undergoing clinical trials as a candidate agent for a vaccine against this species of parasite (Yazdani et al. 2004 ). The African Duffy-negative allele, denoted “FY*O,” has the highest F ST value observed in humans and has other features that are strongly suggestive of recent positive selection (Hamblin and Di Rienzo 2000 ; Hamblin et al. 2002 ). There is further support for selective pressure at this locus, from the observation of an entirely independent FY polymorphism, which has emerged in Papua New Guinea, that decreases Duffy-antigen expression and acts to reduce P. vivax invasion efficiency (Zimmerman et al. 1999 ; Michon et al. 2001 ). The apparent strength of selection at the FY locus is somewhat puzzling for malariologists, since P. vivax infection is not generally lethal, and it has even been proposed that P. vivax infection may protect against the much more lethal parasite P. falciparum (Williams et al. 1996 ).
The study of human genetic polymorphisms has also been informative about how P. falciparum invades erythrocytes, but, in contrast to P. vivax , the available data suggest multiple invasion pathways with considerable redundancy (Hadley et al. 1987 ). A lot of attention has focused on GYPA, GYPB, and GYPC, the genes encoding glycophorins A, B, and C, respectively.
Various blood groups are determined by the erythrocyte-membrane sialoglycoproteins glycophorin A and B, and genetic deficiency of glycophorin A or B expression makes erythrocytes relatively resistant to invasion by P. falciparum (Facer 1983 ). Specific sialic-acid residues on the glycophorin A molecule are recognized by a Duffy-binding–like domain of P. falciparum erythrocyte-binding antigen 175 (Orlandi et al. 1992 ; Mayor et al. 2005 ). Sequence analysis shows evidence of strong evolutionary selection, not only for GYPA and GYPB in the human host (Baum et al. 2002 ; Wang et al. 2003 ), but also for EBA-175 in the P. falciparum parasite, which implies an ongoing evolutionary struggle between the parasite ligand and the host receptor (Wang et al. 2003 ).
Glycophorin C is a minor component of the erythrocyte membrane that serves as a receptor for the P. falciparum erythrocyte-binding antigen 140 (EBA140). The Gerbich-negative blood group, caused by a deletion of GYPC exon 3, is common in coastal areas of Papua New Guinea and results in reduced invasion by P. falciparum (Maier et al. 2003 ). Epidemiological studies indicate that this GYPC deletion does not alter the prevalence or density of asymptomatic malaria infection, but so far there has been no study of how it affects the clinical severity of infection (Patel et al. 2001 , 2004 ).
Another erythrocyte-membrane protein that has been implicated in malaria resistance is an anion exchanger known as “band 3 protein,” encoded by SLC4A1. A 27-bp deletion in this gene results in a form of ovalocytosis that is common in parts of Southeast Asia. It appears to be protective both against malaria infection (Cattani et al. 1987 ; Foo et al. 1992 ) and against cerebral malaria (Genton et al. 1995 ; Allen et al. 1999 ). The mechanism of protection is not yet known. It may relate to the involvement of band 3 protein in endothelial cytoadherence or to some inhibitory effect on parasite invasion or growth.
Structural Variation of Globin Genes
Erythrocytes are essentially bags filled with hemoglobin, and the malaria parasite has developed a lifestyle that is hugely dependent on its hemoglobin environment. Alterations in hemoglobin may affect the biochemical and cellular machinery of parasite development, and they may affect the ability of the spleen and other immune mechanisms to recognize parasites, by affecting the morphology, mechanical properties, or surface structure of the parasitized erythrocyte. The biological importance of these dependencies is highlighted by the huge selective pressure that malaria has exerted on the structure and regulation of α globin (encoded by the identical HBA1 and HBA2 genes) and β globin (encoded by HBB ) that together comprise the tetrameric protein backbone of adult hemoglobin.
HBB has three different coding SNPs that each confer resistance against malaria and that have risen to high frequency in different populations. The HbS allele is a glutamic acid→valine substitution at codon 6 of the β globin chain, HbC is a glutamine→lysine substitution at codon 6, and HbE allele is a glutamic acid→lysine substitution at codon 26. The corresponding proteins are known as “hemoglobin S” (or “sickle hemoglobin”), “hemoglobin C,” and “hemoglobin E.”
The HbS allele is found across a large part of sub-Saharan African as well as parts of the Middle East. It has the distinction of being one of the first human genetic variants to be associated with a specific molecular defect (Pauling et al. 1949 ). Hemoglobin S tends to polymerize at low oxygen concentrations, which causes the erythrocyte to deform into a sickle-like shape (Brittenham et al. 1985 ). HbS homozygotes have sickle-cell disease, a debilitating and often fatal disorder caused by these red-cell deformities. The heterozygous state (denoted “HbAS”) is not generally associated with any clinical abnormality and confers ∼10-fold increase in protection from life-threatening forms of malaria, with a lesser degree of protection against milder forms of the disease (Allison 1954 ; Gilles et al. 1967 ; Hill et al. 1991 ; Allen et al. 1992 ; Stirnadel et al. 1999 b ; Sokhna et al. 2000 ; Ackerman et al. 2005 ). It is still not known precisely how HbAS protects against malaria. Two plausible mechanisms, which are not mutually exclusive, are suppression of parasite growth in red cells (Pasvol et al. 1978 ) and enhanced splenic clearance of parasitized erythrocytes (Shear et al. 1993 ). A study of Kenyan children found that the protective effect of HbAS against malaria increased from 20% to 56% between the ages of 2 and 10 years, which implies that it enhances or acts in synergy with the acquired immune response (Williams et al. 2005 a ). The trade-off between risks and benefits acts to maintain the HbS polymorphism at allele frequencies of ∼10% in many parts of Africa, despite the lethal consequences for homozygotes, which provides the most striking known example of heterozygote advantage in human genetics.
Hemoglobin C is found in several parts of West Africa, although less commonly than is HbS. It results in a much-less-damaging clinical phenotype than sickle-cell disease: homozygotes have a relatively mild hemolytic anemia, and heterozygotes do not experience a significant reduction in hemoglobin levels (Diallo et al. 2004 ). Both heterozygotes and homozygotes of HbC are protected against severe malaria (Agarwal et al. 2000 ; Mockenhaupt et al. 2004 a ; Rihet et al. 2004 ), but the protective effect appears to be substantially greater in homozygotes (Modiano et al. 2001 b ). It has been proposed that the protective effect of HbC may operate by increasing immune clearance of infected erythrocytes. This is based on observations of reduced parasite cytoadherence, abnormal PfEMP1 expression, clustering of erythrocyte band 3 protein, and altered surface topography of the erythrocyte membrane in the presence of hemoglobin C (Arie et al. 2005 ; Fairhurst et al. 2005 ; Tokumasu et al. 2005 ).
Hemoglobin E is common in Southeast Asia, with carrier rates of 50% in some places, and analysis of haplotype structure suggests that the mutation is relatively recent and has risen rapidly in allele frequency (Ohashi et al. 2004 ). Homozygotes generally have symptomless anemia. Although it has not been epidemiologically proven that HbE protects against severe malaria, this is assumed to be the case, and it has been observed that erythrocytes from HbE-heterozygous individuals are relatively resistant to invasion by P. falciparum (Chotivanich et al. 2002 ).
Regulatory Variation of Globin Genes
The thalassemias are the most common Mendelian diseases of humans and constitute a major global health problem (Weatherall and Clegg 2001 ). They comprise a group of clinical disorders that result from defective production of α- or β-globin chains, which arise from deletions or other disruptions of the globin gene clusters on chromosomes 11 and 16. There is a broad spectrum of clinical phenotypes, reflecting the range of different genetic variants that exist and given greater complexity by the fact that α-globin is produced by two identical genes, HBA1 and HBA2. Broadly speaking, homozygous thalassemia results in severe disease or is fatal, whereas heterozygotes are healthy apart from mild anemia. An exception to this general rule arises when either the HBA1 or the HBA2 gene, but not both, is disrupted, so that some α-globin production is possible. This condition is known as “α + thalassemia,” and α + thalassemia homozygotes are only mildly anemic.
Over half a century ago, J. B. S. Haldane proposed balanced polymorphism as the explanation for why thalassemia had risen to high frequencies—approaching fixation—in certain populations (Haldane 1949 ). He argued that heterozygotes might be protected against some important disease, and malaria was the obvious candidate, since the global distribution of thalassemia encompasses the major malarious regions of Africa and Asia and Mediterranean regions where malaria was once common. There are many other circumstantial lines of evidence to support this concept; for example, the Tharu people have both a much higher allele frequency of α thalassemia, of ∼0.8, and a much lower incidence of malarial illness than do other ethnic groups that inhabit the same region of Nepal (Modiano et al. 1991 ). Arguably the strongest population-genetic evidence comes from a detailed survey in Melanesia that showed that the frequency of α + thalassemia varied according to both altitude and latitude in a manner that was highly correlated with malaria endemicity, whereas haplotypic analysis seemed to rule out the possibility that this could have arisen because of founder effects (Flint et al. 1986 ).
Although the population-genetic evidence seems overwhelming, it is only relatively recently that direct evidence that thalassemia protects against malaria has emerged, and the case is still not absolutely clear-cut. A study of Kenyan children found that both heterozygous and homozygous α + thalassemia was protective against severe malaria (Williams et al. 2005 b ), whereas a study of Ghanaian children found that heterozygotes were protected (Mockenhaupt et al. 2004 b ). In Papua New Guinea, the risk of severe malaria was found to be reduced by 60% in children who were homozygous for α + thalassemia and to a lesser degree in heterozygotes, but the result did not seem to be malaria-specific, since a protective effect was also observed for other childhood infections (Allen et al. 1997 ).
The protective mechanism of thalassemia is unknown. Flow-cytometry studies in vitro have shown that erythrocytes with the α thalassemia phenotype show reduced parasite growth (Pattanapanyasat et al. 1999 ) and increased binding of antibodies from malaria-immune sera (Williams et al. 2002 ). Enhanced splenic clearance of malaria-infected cells is a further possibility but is difficult to test in vivo. However, much more complex explanations are also possible, as illustrated by findings of data from Vanuatu which are unusual in two respects: first, both P. falciparum and P. vivax infection are common, but severe malaria is remarkably uncommon in this population; and, second, young children with α + thalassemia have a significantly higher incidence of malaria than do nonthalassemic children (Williams et al. 1996 ). This latter result is exactly the opposite of what Williams et al. ( 1996 ) set out to prove, but they came up with a possible explanation that was based on the further observation that P. vivax infection, which does not cause severe disease, is acquired at an earlier age in this population than is P. falciparum infection. The proposal is that, in this particular epidemiological scenario, α + thalassemia may enhance early exposure to P. vivax infection, thereby in some way protecting against severe disease from later exposure to P. falciparum.
Oxidative Stress
Malaria parasites need to break down hemoglobin to make room to grow, quite apart from any nutritional benefit they may derive from this. This process releases by-products that are potentially toxic—particularly iron, which is a source of oxidative stress.
An important form of defense against oxidative stress within the erythrocyte is production of the electron donor nicotinamide adenine dinucleotide phosphate by the enzyme glucose-6-phosphate dehydrogenase (G6PD), encoded by G6PD on chromosome X. There are many different variants of G6PD, and those that markedly compromise enzyme activity result in hemolytic anemia. The geographical distribution of G6PD deficiency is consistent with evolutionary selection by malaria (Ganczakowski et al. 1995 ), and analysis of haplotypic structure at the G6PD locus supports the hypothesis of recent positive selection (Tishkoff et al. 2001 ; Sabeti et al. 2002 b ). Deficient G6PD enzyme activity has been shown to correlate with protection against severe malaria in Nigerian children (Gilles et al. 1967 ). A study of >2,000 Gambian and Kenyan children found that the common African form of G6PD deficiency (G6PD A − ) is associated with ∼50% reduced risk of severe malaria in female heterozygotes and in male hemizygotes (Ruwende et al. 1995 ). Reduced parasite replication in G6PD-deficient erythrocytes is thought to be the mechanism of protection (Luzzatto et al. 1969 ), but the parasite appears to counter this by manufacturing G6PD itself (Usanga and Luzzatto 1985 ).
Haptoglobin, encoded by HP, is not an erythrocyte protein but is mentioned here because it is a hemoglobin-binding protein that is present in plasma. It could act to defend the malaria-infected individual in several ways: by trapping free hemoglobin, it helps to prevent hemoglobin-induced oxidative tissue damage; it has been shown to inhibit the development of malaria parasites in vitro (Imrie et al. 2004 ); and it appears to reduce parasite load, as determined by murine gene knockout studies (Hunt et al. 2001 ). The haptoglobin 1-1 genotype, characterized by protein electrophoresis, has been associated with susceptibility to severe P. falciparum malaria (Elagib et al. 1998 ; Quaye et al. 2000 ), although a DNA-based study of haptoglobin polymorphisms in the Gambia failed to detect such an association (Aucan et al. 2002 )
Cytoadherence, a Major Factor in Malaria Pathogenesis
A critical event in the pathogenesis of severe malaria is the sequestration of P. falciparum –infected erythrocytes in small blood vessels (Taylor et al. 2004 ). A range of receptor-ligand interactions causes parasitized erythrocytes to stick to endothelium, platelets, and other erythrocytes; this is thought to be an immune-evasion strategy that allows the parasite to stay within the vascular compartment but to avoid circulating through the spleen. On the parasite side, the major ligand is P. falciparum erythrocyte-membrane protein-1 (PfEMP-1)—encoded by a gene family called “ var ” because each parasite contains many different copies of the gene—and, by switching expression between the copies, it is able to cause a remarkable degree of antigenic variation (Kyes et al. 2001 ). On the host side, a range of different adhesion molecules expressed on endothelium, platelets, macrophages, and other erythrocytes serves as binding receptors for different forms of PfEMP-1.
A number of associations have been reported between severe malaria and polymorphisms of host receptors for cytoadherence by P. falciparum –infected erythrocytes ( table 2 ). When the same polymorphism has been tested in different geographical locations, the results have been variable—not simply failure to replicate but, in some cases, the association of the same polymorphism with susceptibility to severe malaria in one study and with resistance in another. As with all genetic association studies, it is possible that these results are statistical artifacts that will ultimately be resolved by larger sample sizes and by finer-scale genetic mapping of these loci. However, in this particular area of genetic analysis, it is not out of the question that the functional consequences of a single polymorphism could vary between locations and might even vary over time at a single location. As outlined above, the biological phenomenon of parasite cytoadherence to endothelium and to other cells is driven by the parasite, not by the host. The parasite varies its pattern of sequestration in different organs by constantly switching between different forms of PfEMP-1 that bind to different host receptors (and different parts of the same receptor) in a promiscuous and opportunistic manner (Roberts et al. 1992 ).
Host Molecules That Mediate Cytoadherence by P. falciparum –Infected Erythrocytes and That Have Been Reported to Show Association with Resistance or Susceptibility to Malaria
PE = parasitized erythrocyte.
Many isolates of P. falciparum bind to endothelium via the CD36 antigen (Barnwell et al. 1989 ). Encoded by CD36, this is a receptor for a range of different molecules, including thrombospondin and long-chain fatty acids, and is expressed by platelets and dendritic cells as well as endothelium. It is a molecule of considerable interest to malaria immunologists because, as well as being a mediator of parasite sequestration, CD36 acts to bind parasitized erythrocytes to dendritic cells, an event that seems to incapacitate the dendritic cell when it comes to presenting parasite antigens (Urban et al. 1999 , 2001 ). Several CD36 polymorphisms have been described in malarious regions (Aitman et al. 2000 ; Omi et al. 2003 ), but the results of disease-association studies are confusing. A study of both Gambian and Kenyan case-control samples found that homozygotes for a nonsense polymorphism, the CD36 + 1264G allele, were susceptible to cerebral malaria (Aitman et al. 2000 ), but a study of the same allele in Kenya alone found that heterozygosity was associated with protection against severe malaria (Pain et al. 2001 ). In Thailand, a dinucleotide repeat sequence in intron 3, implicated in alternative splicing, has been associated with protection against cerebral malaria (Omi et al. 2003 ).
Some isolates of P. falciparum bind strongly to endothelium via intercellular adhesion molecule-1 (Berendt et al. 1989 ). This is encoded by ICAM1, and its normal function is to serve as an endothelial- and immune-cell adhesion receptor for integrin-expressing leukocytes. A polymorphism in the N-terminal domain, present at high frequencies in African populations, acts to reduce binding (Fernandez-Reyes et al. 1997 ). A study in Gabon found that the low-binding allele was associated with reduced susceptibility to severe malaria (Kun et al. 1999 ), but a study in Kenya found increased susceptibility (Fernandez-Reyes et al. 1997 ), and another in the Gambia found no significant effect (Bellamy et al. 1998 a ). A survey of different ethnic groups in India found a number of novel ICAM1 variants that merit analysis in disease-association studies (Sengupta et al. 2004 ).
Another endothelial-binding receptor for P. falciparum is the platelet–endothelial cell adhesion molecule, encoded by PECAM1 (Treutiger et al. 1997 ). A common coding variant (Leu→Val at codon 125) was analyzed in case-control studies of severe malaria in Papua New Guinea and Kenya, but no significant association was identified (Casals-Pascual et al. 2001 ). A study from Thailand has reported a PECAM1 haplotype that is more common in cerebral malaria than in other forms of severe malaria (Kikuchi et al. 2001 ).
Parasite sequestration is not due only to endothelial binding. Some P. falciparum isolates show a phenomenon known as rosetting, where a parasitized erythrocyte binds to other erythrocytes. One mechanism for rosetting is through PfEMP-1 binding to erythrocyte–complement receptor 1, encoded by CR1 (Rowe et al. 1997 ). Up to 80% of the population of a malarious region of Papua New Guinea have erythrocyte CR1 deficiency, which has been associated both with polymorphisms in the CR1 gene and with α + thalassemia, which is also common in this population (Cockburn et al. 2004 ). In the same study, it was found that both CR1 polymorphisms and α thalassemia were independently associated with resistance to severe malaria. However, studies of CR1 polymorphisms in the Gambia found no evidence of association with disease severity (Bellamy et al. 1998 a ; Zimmerman et al. 2003 ). In Thailand, an RFLP that is associated with reduced expression of CR1 on erythrocytes has been associated (in homozygotes) with susceptibility to severe malaria (Nagayasu et al. 2001 ).
Malaria and the Immune System
The genetic interaction between malaria and the immune system is potentially of huge practical interest, for two distinct reasons. First, although there is a vast literature on immunological responses to malaria in humans and in experimental model systems, there is still surprisingly little concrete evidence about precisely which immunological responses are causal mechanisms of protective immunity in naturally exposed populations, and this is a fundamental roadblock in the development of an effective malaria vaccine. One way to establish causality is to obtain clear-cut evidence that functional variation in a specific immune gene affects the clinical outcome of infection. Second, just as the selective pressure of malaria on the erythrocyte has led to common hematological disorders, such as sickle-cell disease and thalassemia, it is possible that we might learn a great deal about mechanisms of chronic immunological and inflammatory disorders if we had a better understanding of the selective pressure that malaria has exerted on the immune system.
Here, we consider a number of immune gene associations with malaria resistance and susceptibility that have emerged over the past 15 years ( table 3 ). The same caveats, discussed above for adhesion molecules, apply here. Few of these associations have been tested in several different studies and, when this has been done, the results have been variable. Thus, much of this section should be regarded as indicative of ongoing research activity rather than definitive results. However, it is important to bear in mind that there potentially are biological reasons why the same immune gene polymorphism might have different consequences in different malarious regions. For example, HLA associations might vary according to the local prevalence of critical parasite-antigen polymorphisms. And associations with inflammatory cytokines and other immune genes may be affected by regional differences in the intensity of malaria transmission, which have complex consequences for the development of acquired immunity and the pattern of severe disease (Snow et al. 1997 ). These are issues that may have far-reaching implications for vaccine efficacy, and, to tackle them robustly, we need much larger sample sizes and more fine-grained genetic association maps than we have at present.
Immune Genes Reported to Be Associated with Different Malaria Phenotypes
Antigen Recognition
In malaria, the opportunities for antigen presentation are limited by the fact that erythrocytes do not express MHC molecules. Liver cells, however, express MHC class I molecules and therefore provide a potential target for cytotoxic T cell (CTL) responses during the first phase of malaria infection, when parasites replicate in the liver prior to invading erythrocytes. HLA-B is an exceptionally polymorphic gene that encodes an MHC class I heavy chain that, together with β2 microglobulin, makes up the HLA-B antigen–presentation complex. The HLA-B53 allele is extremely common in West Africa, compared with other parts of the world, and is associated with a significantly reduced risk of severe malaria in Gambian children (Hill et al. 1991 ). In view of the fact that HLA-B is expressed by liver cells but not by erythrocytes, this genetic association implies that liver-stage parasites provide a significant target for naturally acquired protective immunity. This has boosted efforts to develop a liver-stage malaria vaccine, and it has been proposed that T cell epitope targets for malaria-vaccine development may be obtained by analyzing the peptides that bind to HLA-B53 (Hill et al. 1992 ).
But when a polymorphic parasite antigen interacts with a polymorphic host antigen–presenting system, there are many opportunities for complexity. For example, within a single fragment of the P. falciparum circumsporozoite protein (CSP), it has been observed that the Gambian parasite population has two different variants (cp26 and cp29) that bind to HLA-B53, and these two variants are found together in the same individual more frequently than expected by chance. Although both cp26 and cp29 are effective targets for CTLs at the individual level, they appear to show immunological antagonism such that each acts to suppress the CTL response to the other, and it has been postulated that this is an immune-evasion strategy on the part of the parasite (Gilbert et al. 1998 ).
HLA-DRB1 encodes an HLA class II β chain that, together with an α chain, comprise the HLA-DR antigen–presenting complex. This is expressed in B lymphocytes, dendritic cells, and macrophages and is crucial for antibody production. HLA DRB1*1302-DQB1*0501 has been associated with resistance to severe malaria in Gambian children (Hill et al. 1991 ), and the incidence of malaria-fever episodes in Gambian children is reported to show an overall association with distribution of MHC class II haplotypes (Bennett et al. 1993 ).
The Antibody Response
Interleukin-4, encoded by IL4, is produced by activated T cells and promotes proliferation and differentiation of antibody-producing B cells. A study of the Fulani of Burkina Faso, who have both fewer malaria attacks and higher levels of antimalarial antibodies than do neighboring ethnic groups, found that the IL4 -524 T allele was associated with elevated antibody levels against malaria antigens, which raises the possibility that this might be a factor in increased resistance to malaria (Luoni et al. 2001 ).
CD40 ligand, encoded by the X chromosome gene TNFSF5, is expressed in T cells and binds to CD40 in B cells, which acts to regulate immunoglobulin class switching and other aspects of B cell function. In a Gambian case-control study, the TNFSF5 -726C allele was associated with protection against severe malaria (Sabeti et al. 2002 a ), and long-range haplotype analysis of this allele suggests that it has recently undergone positive evolutionary selection (Sabeti et al. 2002 b ).
Many leukocytes express receptors for the Fc portion of IgG, which are used to engage and remove antigen-antibody complexes. A His→Arg substitution at codon 131 of FCGR2A, which encodes low-affinity IIa receptor for the Fc fragment of IgG, results in failure to bind to IgG2 and has been associated with protection against high levels of P. falciparum parasitemia in Kenya (Shi et al. 2001 ). Follow-up studies in Thailand and the Gambia found that homozygotes for the 131His genotype are susceptible to cerebral malaria (Omi et al. 2002 ; Cooke et al. 2003 ). In the Thai study, the FCGR2A association involved interaction with an FCGR3B gene polymorphism.
The Proinflammatory Response
Tumor necrosis factor is encoded by the TNF gene located in the MHC class III region, flanked by the MHC class I and II regions. It is a proinflammatory cytokine that is critical for innate immunity against malaria parasites but has also been implicated in the pathogenesis of severe malaria (reviewed by Kwiatkowski [ 1995 ] and Clark et al. [ 2004 ]). Two longitudinal family studies, in the Gambia and Burkina Faso, have identified linkage between the MHC region and susceptibility to malaria-fever episodes, and TNF is at the center of the linkage peaks (Jepson et al. 1997 ; Flori et al. 2003 ). Several TNF-promoter polymorphisms have been independently associated with severe malaria. Gambian children who are homozygous for the TNF -308A allele have been observed to have increased susceptibility to cerebral malaria (McGuire et al. 1994 ), and, in Gabon, it was found that those who carried this allele were more likely to encounter symptomatic reinfections with P. falciparum (Meyer et al. 2002 ). A study in Sri Lanka found that the carriers of the TNF -308A allele had increased risk of severe infectious diseases in general (Wattavidanage et al. 1999 ), whereas a study in Kenya found an increase in infant mortality and malaria morbidity (Aidoo et al. 2001 ). TNF -376A confers allele-specific binding of the transcription factor OCT-1 and has been associated with susceptibility to cerebral malaria (Knight et al. 1999 ), whereas the TNF -238A allele has been associated with susceptibility to severe malarial anemia (McGuire et al. 1999 ). A longitudinal study in Burkina Faso suggests that several different TNF-promoter SNPs are involved in the regulation of parasite density (Flori et al. 2005 ). The functional role of TNF -308 and other TNF polymorphisms remains open to question (Abraham and Kroeger 1999 ; Knight et al. 2003 ; Bayley et al. 2004 ), but the surrounding MHC class III region has many other interesting immunological genes and complex patterns of linkage disequilibrium (Ackerman et al. 2003 ). Thus, although TNF is unquestionably an important mediator of both immunity and pathogenesis for malaria, it remains possible that the observed genetic associations with TNF polymorphisms arise from functional variation in neighboring genes.
Inducible nitric oxide (NO) synthase, encoded by NOS2A, generates NO. This is a free radical, with antiparasitic properties, but it also has a potential immunosuppressive role and has been proposed as a factor in cerebral malaria because of its role in neurotransmission (reviewed by Clark and Rockett [ 1996 ]). The NOS2A -954C allele has been associated with elevated NO synthase activity in cells from Gabonese individuals, and, in that population, it has been associated with protection from severe malaria and resistance to reinfection (Kun et al. 1998 , 2001 ), but studies in the Gambia and Tanzania failed to detect such a disease association (Levesque et al. 1999 ; Burgner et al. 2003 ). The NOS2A -1173T allele—which appears, on the basis of measurements in urine and plasma, to be associated with high NO production in Tanzanian children—is associated with protection from malarial illness in Tanzania and from severe malarial anemia in Kenya (Hobbs et al. 2002 ), but no protective effect against severe malaria was detected in the Gambia (Burgner et al. 2003 ). In Gambian children, an NOS2A microsatellite polymorphism has been associated with susceptibility to fatal malaria (Burgner et al. 1998 ), and a haplotype uniquely defined by the NOS2A -1659T allele was associated with cerebral malaria by both the transmission/disequilibrium test (TDT) and case-control analysis (Burgner et al. 2003 ).
Interferon-γ, encoded by IFNG, is a key immunological mediator that is believed to play both a protective and a pathological role in malaria (Stevenson and Riley 2004 ). Analysis of SNPs in the region of IFNG and the neighboring IL22 gene found several weak associations with severe malaria in Gambian children but no clear-cut effect (Koch et al. 2005 ). A study of IFNGR1, which encodes the ligand-binding α chain of the interferon-γ receptor, found that in Mandinka, the major Gambian ethnic group, heterozygotes for the IFNGR1-56 polymorphism were protected against cerebral malaria (Koch et al. 2002 ). Reporter-gene analysis suggests that the minor allele acts to reduce levels of IFNGR1 gene expression (Juliger et al. 2003 ).
IFNAR1 encodes interferon α receptor 1, a type I membrane protein that forms one of the two chains of a receptor for interferons α and β. In a murine malaria model, it has been observed that interferonα inhibits parasite development within erythrocytes (Vigario et al. 2001 ). A Gambian case-control study found two IFNAR1 SNPs that were associated with protection against severe malaria, and a resistance haplotype was identified (Aucan et al. 2003 ).
IL12B encodes a subunit of interleukin-12, a cytokine produced by activated macrophages that is essential for the development of Th1 cells. Homozygotes for an IL12B -promoter polymorphism were found to have decreased NO production when measured in blood samples, and this genotype has been associated with a fatal outcome in cerebral malaria in Tanzanian but not in Kenyan children (Morahan et al. 2002 )
The interleukin-1 family of cytokines, produced mainly by macrophages, are important mediators of the inflammatory response to infection and of fever. In a Gambian case-control study, a SNP in IL1A (encoding interleukin-1α) and another in IL1B (encoding interleukin-1β) showed a marginal association with susceptibility to malaria (Walley et al. 2004 ).
Interleukin-10, encoded by IL10, is a crucial anti-inflammatory cytokine. Several lines of evidence indicate that IL10 is protective against severe malaria and that IL10 production is genetically determined. An analysis of IL10 SNPs in Gambian children found a common haplotype that was strongly associated with protection against severe malaria by case-control analysis but not by TDT analysis of the same population (Wilson et al. 2005 ). Since the case-control analysis was ethnically matched, this raises the question of whether IL10 associations with severe malaria might be confounded by fetal survival rates or other sources of transmission bias, since genetic variation at the IL10 locus has been implicated as a determinant of fertility (Westendorp et al. 2001 ).
Other Serum Factors
MBL2 encodes a serum mannose-binding lectin (MBL) protein that recognizes mannose and N-acetylglucosamine on bacterial pathogens and can activate the classic complement pathway. MBL2 polymorphisms have been associated with susceptibility to various infectious diseases. A study in Gabon found that children with severe malaria had low serum MBL levels compared with those of children with mild malaria and that mutations in codons 54 and 57 of MBL2 (which lead to low protein levels) were present at a higher frequency in those with severe malaria (Luty et al. 1998 ). However, a study in the Gambia failed to replicate this association with severe malaria (Bellamy et al. 1998 b ).
Differences in Resistance to Infection among Inbred Mouse Strains
Inbred strains of mice show marked and consistent differences in their response to malaria infection (Greenberg et al. 1954 ; Greenberg and Kendrick 1959 ; Rest 1982 ; Stevenson et al. 1982 ; Stevenson and Skamene 1985 ). Put simply, some mouse strains are more resistant to malaria than are others, but the details are somewhat more complex, since mice that are relatively resistant to one parasite strain may be relatively susceptible to a different parasite strain. For example, some strains of mice are resistant to nonlethal strains but highly susceptible to lethal strains of Plasmodium yoelii, whereas other mouse strains show the opposite pattern (Sayles and Wassom 1988 ). This confirms that both host and parasite genes—and the specific way in which they are combined—are important determinants of resistance or susceptibility to malaria.
Plasmodium chabaudi Infection: A Model of How the Parasite Population Is Controlled
P. chabaudi infection is controlled and cleared by some inbred strains of mice (e.g., C57BL/6J, C57L/J, DBA/2J, CBA/J, and B10.A/SgSn) much more effectively than by others (e.g., A/J, DBA/1J, BALB/c, C3H/HeJ, AKR/J, and SJL/J) (Stevenson et al. 1982 ). At least some of these interstrain differences show classic Mendelian patterns of segregation in crossbreeding experiments and that show resistance is generally dominant over susceptibility. When susceptible A/J and resistant C57BL mice were crossbred, it was found that the degree of splenic enlargement after infection was genetically linked to the ability to suppress parasitemia (Stevenson and Skamene 1985 ).
Genomewide linkage screens have been performed after crossbreeding susceptible mice (C3H, SJL, or A/J) with resistant mice (C57BL/6J). A locus on chromosome 9 (Char1) determines death or survival (Foote et al. 1997 ). A well-characterized locus on chromosome 8 (Char2) determines the control of parasite density (Foote et al. 1997 ; Fortin et al. 1997 ; Burt et al. 2002 ). A locus in the MHC region of chromosome 17 (Char3) influences parasite clearance rates at the time immediately after peak parasitemia (Burt et al. 1999 ).
A further resistance locus (Char4) was identified after deriving recombinant congenic strains from susceptible A/J and resistant C57BL/6J mice. The Char4 locus maps to a small congenic B6 fragment on chromosome 3 (Fortin et al. 2001 ). Sequencing of candidate genes across this region has identified a plausible functional mutation—namely, a loss-of-function coding variant of the pyruvate kinase gene ( Pklr ). In uninfected animals, this mutation causes hemolytic anemia that is compensated by constitutive reticulocytosis and splenomegaly (Min-Oo et al. 2003 , 2004 ). It is possible that the malaria-protective effect is a direct consequence of impaired viability and increasing splenic clearance of host erythrocytes.
An advanced intercross line population derived from susceptible A/J and resistant C57BL/6J mice was used to identify QTLs for control of parasitemia (Hernandez-Valladares et al. 2004 a ). Of particular interest was a novel QTL (Char 8) in the chromosome 11 region that is homologous to the human 5q31-q33 region discussed above, which is rich in Th2 cytokine genes (Hernandez-Valladares et al. 2004 b ).

Plasmodium berghei ANKA Infection: A Model of Inflammatory Pathology
The ANKA strain of P. berghei causes more-severe pathology than do most other experimental murine parasites. A curious feature of P. berghei ANKA (PBA) infection in some inbred mouse strains (e.g., A/J and C57BL/6) is that 100% of the mice die after 5–8 d, with cerebral hemorrhages as a terminal event (Rest 1982 ). Cloned lines of PBA differ in their tendency to cause these cerebral changes, which indicates that pathology is determined by a specific combination of host and parasite genotype (Amani et al. 1998 ).
PBA-induced cerebral pathology is not a reliable model of human cerebral malaria. In particular, parasite sequestration in cerebral capillaries, a hallmark of human cerebral malaria (Taylor et al. 2004 ), is notable by its absence in the PBA experimental model. The pathological features of PBA in susceptible mouse strains include mononuclear cell adhesion to endothelium, which is absent in human cerebral malaria, together with hemorrhage and cerebral endothelial cell damage, with breakdown of the blood-brain barrier and cerebral edema (Thumwood et al. 1988 ; Neill and Hunt 1992 ; Neill et al. 1993 ). However, PBA has provided an interesting experimental system in which to study how immunopathological processes are affected by different interventions (e.g., Grau et al. [ 1987 a , 1987 b , 1989 ], Kremsner et al. [ 1991 ], Hunt et al. [ 1993 ], Engwerda et al. [ 2002 ], and Schofield et al. [ 2002 ]).
To search for genetic factors that determine PBA-induced cerebral pathology, one approach has been to enlarge the pool of genetic diversity by deriving new inbred mouse strains from wild-mouse populations (Bagot et al. 2002 b ). When a resistant wild-derived inbred strain (WLA) was crossed with a susceptible laboratory strain (C57BL/6J), all of the F 1 progeny and 97% of the F 2 progeny displayed resistance. A genomewide screen, performed after backcrossing the resistant wild strain onto the susceptible laboratory strain, found that resistance was linked to loci on chromosome 1 (Berr1) and chromosome 11 (Berr2) (Bagot et al. 2002 a ).
Recent analysis of the F 2 progeny of WLA and C57BL/6 strains has revealed a fascinating combinatorial effect: it seems that the WLA allele at the Berr1 locus confers resistance to early death from cerebral pathology, whereas the C57Bl/6 allele at a locus on chromosome 9 (Berr3) increases the ability of the mouse to clear the infection. Thus, the progeny have greater resistance to malaria than either of the parental strains (Campino et al. 2005 ).
A study that crossbred susceptible C57BL/6 mice with resistant DBA/2 mice identified a major resistance locus on chromosome 18 (Nagayasu et al. 2002 ). Another study that crossed susceptible CBA mice with resistant DBA/2 mice identified a susceptibility locus at the MHC region on chromosome 17 (Ohno and Nishimura 2004 ).
Conclusions
As geneticists brace themselves to perform genomewide association analysis of common diseases—a task that is going to require massive investment both in epidemiological infrastructure and in genotyping technology—malaria stands out as a target for which this approach is feasible and potentially of huge importance for disease prevention. In terms of feasibility, malaria is the most powerful known force for recent selection of human genetic variants, so malaria-protective polymorphisms are likely to be at high frequencies in affected populations, and, if recently selected, they may also show strong linkage disequilibrium with neighboring genetic markers.
In terms of practical importance for disease prevention, genetic studies of malaria have already yielded results: the observation that Duffy antigen–negative individuals are resistant to infection with P. vivax was the starting point of a chain of molecular discovery that led to a candidate vaccine against P. vivax that is now undergoing trials. A major impetus for researchers working in this area is the hope that large-scale genomic epidemiology will be a way of getting at basic questions that decades of immunological research have failed to resolve, such as how infected individuals clear parasites from the bloodstream or why malaria causes cerebral complications in some people but not others. The holy grail of this field is to discover novel molecular pathways for protective immunity that will provide critical insights for the development of a vaccine to reduce the massive global burden of disease due to P. falciparum.
Malaria research groups across the world have collected DNA samples and detailed clinical data from thousands of individuals with severe malaria, as well as from parents and population controls. Until recently, the resource has been fragmented, with different groups pursuing relatively small studies of their own samples, but there is a growing impetus to link these independent studies to form a global infrastructure for genomic epidemiology of malaria. One such initiative is Malaria Genomic Epidemiology Network (MalariaGEN), which brings together research groups in 15 different malaria-endemic countries. Large-scale epidemiological studies are needed for the sample sizes required to detect modest effects while testing perhaps a million SNP markers (Risch 2000 ). And once the genome-screening phase has been completed, data from different populations are needed to analyze gene-environment interactions and are needed because haplotypic diversity provides the means to dissect functional polymorphisms from nonfunctional genetic markers.
Finally, it should not be forgotten that, as recently as a century ago, malaria was found in parts of Europe and North America, in addition to its current distribution across most of Africa and large parts of Asia and South America. Sickle-cell disease and thalassemia are two classic examples of how the historical effects of malaria have left an imprint on the pattern of disease in contemporary populations. It remains an open question as to whether any of the immunological, inflammatory, and other chronic diseases that are found in modern societies are, in part, due to the evolutionary pressure that malaria exerted on our ancestors.
Acknowledgments
Funding from the Medical Research Council and the Bill and Melinda Gates Foundation is gratefully acknowledged.
Web Resource
The URL for data presented herein is as follows:
- MalariaGEN, http://www.malariagen.net/
- Abel L, Cot M, Mulder L, Carnevale P, Feingold J (1992) Segregation analysis detects a major gene controlling blood infection levels in human malaria. Am J Hum Genet 50:1308–1317 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Abraham LJ, Kroeger KM (1999) Impact of the −308 TNF promoter polymorphism on the transcriptional regulation of the TNF gene: relevance to disease. J Leukoc Biol 66:562–566 [ DOI ] [ PubMed ] [ Google Scholar ]
- Ackerman H, Usen S, Jallow M, Sisay-Joof F, Pinder M, Kwiatkowski DP (2005) A comparison of case-control and family-based association methods: the example of sickle-cell and malaria. Ann Hum Genet ( http://www.blackwell-synergy.com/doi/pdf/10.1111/j.1529-8817.2005.00180.x?cookieSet=1 ) (electronically published April 5, 2005; accessed June 16, 2005) [ DOI ] [ PubMed ]
- Ackerman HC, Ribas G, Jallow M, Mott R, Neville M, Sisay-Joof F, Pinder M, Campbell RD, Kwiatkowski DP (2003) Complex haplotypic structure of the central MHC region flanking TNF in a West African population. Genes Immun 4:476–486 10.1038/sj.gene.6364008 [ DOI ] [ PubMed ] [ Google Scholar ]
- Agarwal A, Guindo A, Cissoko Y, Taylor JG, Coulibaly D, Kone A, Kayentao K, Djimde A, Plowe CV, Doumbo O, Wellems TE, Diallo D (2000) Hemoglobin C associated with protection from severe malaria in the Dogon of Mali, a West African population with a low prevalence of hemoglobin S. Blood 96:2358–2363 [ PubMed ] [ Google Scholar ]
- Aidoo M, McElroy PD, Kolczak MS, Terlouw DJ, ter Kuile FO, Nahlen B, Lal AA, Udhayakumar V (2001) Tumor necrosis factor-alpha promoter variant 2 (TNF2) is associated with pre-term delivery, infant mortality, and malaria morbidity in western Kenya: Asembo Bay Cohort Project IX. Genet Epidemiol 21:201–211 10.1002/gepi.1029 [ DOI ] [ PubMed ] [ Google Scholar ]
- Aitman TJ, Cooper LD, Norsworthy PJ, Wahid FN, Gray JK, Curtis BR, McKeigue PM, Kwiatkowski D, Greenwood BM, Snow RW, Hill AV, Scott J (2000) Malaria susceptibility and CD36 mutation. Nature 405:1015–1016 10.1038/35016636 [ DOI ] [ PubMed ] [ Google Scholar ]
- Allen SJ, Bennett S, Riley EM, Rowe PA, Jakobsen PH, O’Donnell A, Greenwood BM (1992) Morbidity from malaria and immune responses to defined Plasmodium falciparum antigens in children with sickle cell trait in the Gambia. Trans R Soc Trop Med Hyg 86:494–498 10.1016/0035-9203(92)90083-O [ DOI ] [ PubMed ] [ Google Scholar ]
- Allen SJ, O’Donnell A, Alexander ND, Alpers MP, Peto TEA, Clegg JB, Weatherall DJ (1997) α + -Thalassemia protects children against disease caused by other infections as well as malaria. Proc Natl Acad Sci USA 94:14736–14741 10.1073/pnas.94.26.14736 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Allen SJ, O’Donnell A, Alexander ND, Mgone CS, Peto TE, Clegg JB, Alpers MP, Weatherall DJ (1999) Prevention of cerebral malaria in children in Papua New Guinea by Southeast Asian ovalocytosis band 3. Am J Trop Med Hyg 60:1056–1060 [ DOI ] [ PubMed ] [ Google Scholar ]
- Allison AC (1954) Protection afforded by sickle-cell trait against subtertian malareal infection. Br Med J 4857:290–294 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Amani V, Boubou MI, Pied S, Marussig M, Walliker D, Mazier D, Renia L (1998) Cloned lines of Plasmodium berghei ANKA differ in their abilities to induce experimental cerebral malaria. Infect Immun 66:4093–4099 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Arie T, Fairhurst RM, Brittain NJ, Wellems TE, Dvorak JA (2005) Hemoglobin C modulates the surface topography of Plasmodium falciparum -infected erythrocytes. J Struct Biol 150:163–169 10.1016/j.jsb.2005.02.008 [ DOI ] [ PubMed ] [ Google Scholar ]
- Aucan C, Walley AJ, Greenwood BM, Hill AV (2002) Haptoglobin genotypes are not associated with resistance to severe malaria in the Gambia. Trans R Soc Trop Med Hyg 96:327–328 10.1016/S0035-9203(02)90114-8 [ DOI ] [ PubMed ] [ Google Scholar ]
- Aucan C, Walley AJ, Hennig BJ, Fitness J, Frodsham A, Zhang L, Kwiatkowski D, Hill AV (2003) Interferon-alpha receptor-1 (IFNAR1) variants are associated with protection against cerebral malaria in the Gambia. Genes Immun 4:275–282 10.1038/sj.gene.6363962 [ DOI ] [ PubMed ] [ Google Scholar ]
- Bagot S, Campino S, Penha-Goncalves C, Pied S, Cazenave PA, Holmberg D (2002 a ) Identification of two cerebral malaria resistance loci using an inbred wild-derived mouse strain. Proc Natl Acad Sci USA 99:9919–9923 10.1073/pnas.152215199 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Bagot S, Idrissa Boubou M, Campino S, Behrschmidt C, Gorgette O, Guenet JL, Penha-Goncalves C, Mazier D, Pied S, Cazenave PA (2002 b ) Susceptibility to experimental cerebral malaria induced by Plasmodium berghei ANKA in inbred mouse strains recently derived from wild stock. Infect Immun 70:2049–2056 10.1128/IAI.70.4.2049-2056.2002 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Barnwell JW, Asch AS, Nachman RL, Yamaya M, Aikawa M, Ingravallo P (1989) A human 88-kD membrane glycoprotein (CD36) functions in vitro as a receptor for a cytoadherence ligand on Plasmodium falciparum -infected erythrocytes. J Clin Invest 84:765–772 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Baum J, Ward RH, Conway DJ (2002) Natural selection on the erythrocyte surface. Mol Biol Evol 19:223–229 [ DOI ] [ PubMed ] [ Google Scholar ]
- Bayley JP, Ottenhoff TH, Verweij CL (2004) Is there a future for TNF promoter polymorphisms? Genes Immun 5:315–329 10.1038/sj.gene.6364055 [ DOI ] [ PubMed ] [ Google Scholar ]
- Bellamy R, Kwiatkowski D, Hill AV (1998 a ) Absence of an association between intercellular adhesion molecule 1, complement receptor 1 and interleukin 1 receptor antagonist gene polymorphisms and severe malaria in a West African population. Trans R Soc Trop Med Hyg 92:312–316 10.1016/S0035-9203(98)91026-4 [ DOI ] [ PubMed ] [ Google Scholar ]
- Bellamy R, Ruwende C, McAdam KP, Thursz M, Sumiya M, Summerfield J, Gilbert SC, Corrah T, Kwiatkowski D, Whittle HC, Hill AV (1998 b ) Mannose binding protein deficiency is not associated with malaria, hepatitis B carriage nor tuberculosis in Africans. QJM 91:13–18 10.1093/qjmed/91.1.13 [ DOI ] [ PubMed ] [ Google Scholar ]
- Bennett S, Allen SJ, Olerup O, Jackson DJ, Wheeler JG, Rowe PA, Riley EM, Greenwood BM (1993) Human leucocyte antigen (HLA) and malaria morbidity in a Gambian community. Trans R Soc Trop Med Hyg 87:286–287 10.1016/0035-9203(93)90130-I [ DOI ] [ PubMed ] [ Google Scholar ]
- Berendt AR, Simmons DL, Tansey J, Newbold CI, Marsh K (1989) Intercellular adhesion molecule-1 is an endothelial cell adhesion receptor for Plasmodium falciparum. Nature 341:57–59 10.1038/341057a0 [ DOI ] [ PubMed ] [ Google Scholar ]
- Bienzle U, Ayeni O, Lucas AO, Luzzatto L (1972) Glucose-6-phosphate dehydrogenase and malaria: greater resistance of females heterozygous for enzyme deficiency and of males with non-deficient variant. Lancet 1:107–110 10.1016/S0140-6736(72)90676-9 [ DOI ] [ PubMed ] [ Google Scholar ]
- Brittenham GM, Schechter AN, Noguchi CT (1985) Hemoglobin S polymerization: primary determinant of the hemolytic and clinical severity of the sickling syndromes. Blood 65:183–189 [ PubMed ] [ Google Scholar ]
- Burgner D, Usen S, Rockett K, Jallow M, Ackerman H, Cervino A, Pinder M, Kwiatkowski DP (2003) Nucleotide and haplotypic diversity of the NOS2A promoter region and its relationship to cerebral malaria. Hum Genet 112:379–386 [ DOI ] [ PubMed ] [ Google Scholar ]
- Burgner D, Xu W, Rockett K, Gravenor M, Charles IG, Hill AV, Kwiatkowski D (1998) Inducible nitric oxide synthase polymorphism and fatal cerebral malaria. Lancet 352:1193–1194 10.1016/S0140-6736(05)60531-4 [ DOI ] [ PubMed ] [ Google Scholar ]
- Burt RA, Baldwin TM, Marshall VM, Foote SJ (1999) Temporal expression of an H2-linked locus in host response to mouse malaria. Immunogenetics 50:278–285 10.1007/s002510050603 [ DOI ] [ PubMed ] [ Google Scholar ]
- Burt RA, Marshall VM, Wagglen J, Rodda FR, Senyschen D, Baldwin TM, Buckingham LA, Foote SJ (2002) Mice that are congenic for the char2 locus are susceptible to malaria. Infect Immun 70:4750–4753 10.1128/IAI.70.8.4750-4753.2002 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Campino S, Bagot S, Bergman ML, Almeida P, Sepulveda N, Pied S, Penha-Goncalves C, Holmberg D, Cazenave PA (2005) Genetic control of parasite clearance leads to resistance to Plasmodium berghei ANKA infection and confers immunity. Genes Immun ( http://www.nature.com/gene/journal/vaop/ncurrent/full/6364219a.html ) (electronically published June 23, 2005; accessed July 1, 2005) [ DOI ] [ PubMed ] [ Google Scholar ]
- Casals-Pascual C, Allen S, Allen A, Kai O, Lowe B, Pain A, Roberts DJ (2001) Short report: codon 125 polymorphism of CD31 and susceptibility to malaria. Am J Trop Med Hyg 65:736–737 [ DOI ] [ PubMed ] [ Google Scholar ]
- Cattani JA, Gibson FD, Alpers MP, Crane GG (1987) Hereditary ovalocytosis and reduced susceptibility to malaria in Papua New Guinea. Trans R Soc Trop Med Hyg 81:705–709 10.1016/0035-9203(87)90001-0 [ DOI ] [ PubMed ] [ Google Scholar ]
- Chebloune Y, Pagnier J, Trabuchet G, Faure C, Verdier G, Labie D, Nigon V (1988) Structural analysis of the 5′ flanking region of the beta-globin gene in African sickle cell anemia patients: further evidence for three origins of the sickle cell mutation in Africa. Proc Natl Acad Sci USA 85:4431–4435 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Chitnis CE, Miller LH (1994) Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J Exp Med 180:497–506 10.1084/jem.180.2.497 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Chotivanich K, Udomsangpetch R, Pattanapanyasat K, Chierakul W, Simpson J, Looareesuwan S, White N (2002) Hemoglobin E: a balanced polymorphism protective against high parasitemias and thus severe P. falciparum malaria. Blood 100:1172–1176 [ PubMed ] [ Google Scholar ]
- Clark IA, Alleva LM, Mills AC, Cowden WB (2004) Pathogenesis of malaria and clinically similar conditions. Clin Microbiol Rev 17:509–539 10.1128/CMR.17.3.509-539.2004 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Clark IA, Rockett KA (1996) Nitric oxide and parasitic disease. Adv Parasitol 37:1–56 [ DOI ] [ PubMed ] [ Google Scholar ]
- Cockburn IA, Mackinnon MJ, O’Donnell A, Allen SJ, Moulds JM, Baisor M, Bockarie M, Reeder JC, Rowe JA (2004) A human complement receptor 1 polymorphism that reduces Plasmodium falciparum rosetting confers protection against severe malaria. Proc Natl Acad Sci USA 101:272–277 10.1073/pnas.0305306101 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Coluzzi M (1999) The clay feet of the malaria giant and its African roots: hypotheses and inferences about origin, spread and control of Plasmodium falciparum. Parassitologia 41:277–283 [ PubMed ] [ Google Scholar ]
- Coluzzi M, Sabatini A, della Torre A, Di Deco MA, Petrarca V (2002) A polytene chromosome analysis of the Anopheles gambiae species complex. Science 298:1415–1418 10.1126/science.1077769 [ DOI ] [ PubMed ] [ Google Scholar ]
- Conway DJ (2003) Tracing the dawn of Plasmodium falciparum with mitochondrial genome sequences. Trends Genet 19:671–674 10.1016/j.tig.2003.10.007 [ DOI ] [ PubMed ] [ Google Scholar ]
- Cooke GS, Aucan C, Walley AJ, Segal S, Greenwood BM, Kwiatkowski DP, Hill AV (2003) Association of Fcgamma receptor IIa (CD32) polymorphism with severe malaria in West Africa. Am J Trop Med Hyg 69:565–568 [ PubMed ] [ Google Scholar ]
- Cot M, Abel L, Roisin A, Barro D, Yada A, Carnevale P, Feingold J (1993) Risk factors of malaria infection during pregnancy in Burkina Faso: suggestion of a genetic influence. Am J Trop Med Hyg 48:358–364 [ DOI ] [ PubMed ] [ Google Scholar ]
- Diallo DA, Doumbo OK, Dicko A, Guindo A, Coulibaly D, Kayentao K, Djimde AA, Thera MA, Fairhurst RM, Plowe CV, Wellems TE (2004) A comparison of anemia in hemoglobin C and normal hemoglobin A children with Plasmo dium falciparum malaria. Acta Trop 90:295–299 10.1016/j.actatropica.2004.02.005 [ DOI ] [ PubMed ] [ Google Scholar ]
- Dolo A, Modiano D, Maiga B, Daou M, Dolo G, Guindo H, Ba M, Maiga H, Coulibaly D, Perlman H, Blomberg MT, Toure YT, Coluzzi M, Doumbo O (2005) Difference in susceptibility to malaria between two sympatric ethnic groups in Mali. Am J Trop Med Hyg 72:243–248 [ PubMed ] [ Google Scholar ]
- Elagib AA, Kider AO, Akerstrom B, Elbashir MI (1998) Association of the haptoglobin phenotype (1-1) with falciparum malaria in Sudan. Trans R Soc Trop Med Hyg 92:309–311 10.1016/S0035-9203(98)91025-2 [ DOI ] [ PubMed ] [ Google Scholar ]
- Engwerda CR, Mynott TL, Sawhney S, De Souza JB, Bickle QD, Kaye PM (2002) Locally up-regulated lymphotoxin alpha, not systemic tumor necrosis factor alpha, is the principle mediator of murine cerebral malaria. J Exp Med 195:1371–1377 10.1084/jem.20020128 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Facer CA (1983) Merozoites of P. falciparum require glycophorin for invasion into red cells. Bull Soc Pathol Exot Filiales 76:463–469 [ PubMed ] [ Google Scholar ]
- Fairhurst RM, Baruch DI, Brittain NJ, Ostera GR, Wallach JS, Hoang HL, Hayton K, Guindo A, Makobongo MO, Schwartz OM, Tounkara A, Doumbo OK, Diallo DA, Fujioka H, Ho M, Wellems TE (2005) Abnormal display of PfEMP-1 on erythrocytes carrying haemoglobin C may protect against malaria. Nature 435:1117–1121 10.1038/nature03631 [ DOI ] [ PubMed ] [ Google Scholar ]
- Farouk SE, Dolo A, Bereczky S, Kouriba B, Maiga B, Farnert A, Perlmann H, Hayano M, Montgomery SM, Doumbo OK, Troye-Blomberg M (2005) Different antibody- and cytokine-mediated responses to Plasmodium falciparum parasite in two sympatric ethnic tribes living in Mali. Microbes Infect 7:110–117 10.1016/j.micinf.2004.09.012 [ DOI ] [ PubMed ] [ Google Scholar ]
- Feng Z, Smith DL, McKenzie FE, Levin SA (2004) Coupling ecology and evolution: malaria and the S-gene across time scales. Math Biosci 189:1–19 10.1016/j.mbs.2004.01.005 [ DOI ] [ PubMed ] [ Google Scholar ]
- Fernandez-Reyes D, Craig AG, Kyes SA, Peshu N, Snow RW, Berendt AR, Marsh K, Newbold CI (1997) A high frequency African coding polymorphism in the N-terminal domain of ICAM-1 predisposing to cerebral malaria in Kenya. Hum Mol Genet 6:1357–1360 10.1093/hmg/6.8.1357 [ DOI ] [ PubMed ] [ Google Scholar ]
- Flint J, Harding RM, Boyce AJ, Clegg JB (1998) The population genetics of the haemoglobinopathies. Baillieres Clin Haematol 11:1–51 [ DOI ] [ PubMed ] [ Google Scholar ]
- Flint J, Hill AVS, Bowden DK, Oppenheimer SJ, Sill PR, Serjeantson SW, Bana-Koiri J, Bhatia K, Alpers MP, Boyce AJ, Weatherall DJ, Clegg JB (1986) High frequencies of alpha-thalassaemia are the result of natural selection by malaria. Nature 321:744–750 10.1038/321744a0 [ DOI ] [ PubMed ] [ Google Scholar ]
- Flori L, Delahaye NF, Iraqi FA, Hernandez-Valladares M, Fumoux F, Rihet P (2005) TNF as a malaria candidate gene: polymorphism-screening and family-based association analysis of mild malaria attack and parasitemia in Burkina Faso. Genes Immun ( http://www.nature.com/gene/journal/vaop/ncurrent/full/6364231a.html ) (electronically published June 2, 2005; accessed June 16, 2005) [ DOI ] [ PubMed ] [ Google Scholar ]
- Flori L, Sawadogo S, Esnault C, Delahaye NF, Fumoux F, Rihet P (2003) Linkage of mild malaria to the major histocompatibility complex in families living in Burkina Faso. Hum Mol Genet 12:375–378 10.1093/hmg/ddg033 [ DOI ] [ PubMed ] [ Google Scholar ]
- Foo LC, Rekhraj V, Chiang GL, Mak JW (1992) Ovalocytosis protects against severe malaria parasitemia in the Malayan aborigines. Am J Trop Med Hyg 47:271–275 [ DOI ] [ PubMed ] [ Google Scholar ]
- Foote SJ, Burt RA, Baldwin TM, Presente A, Roberts AW, Laural YL, Lew AM, Marshall VM (1997) Mouse loci for malaria-induced mortality and the control of parasitaemia. Nat Genet 17:380–381 10.1038/ng1297-380 [ DOI ] [ PubMed ] [ Google Scholar ]
- Fortin A, Belouchi A, Tam MF, Cardon L, Skamene E, Stevenson MM, Gros P (1997) Genetic control of blood parasitaemia in mouse malaria maps to chromosome 8. Nat Genet 17:382–383 10.1038/ng1297-382 [ DOI ] [ PubMed ] [ Google Scholar ]
- Fortin A, Cardon LR, Tam M, Skamene E, Stevenson MM, Gros P (2001) Identification of a new malaria susceptibility locus (Char4) in recombinant congenic strains of mice. Proc Natl Acad Sci USA 98:10793–10798 10.1073/pnas.191288998 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Ganczakowski M, Town M, Bowden DK, Vulliamy TJ, Kaneko A, Clegg JB, Weatherall DJ, Luzzatto L (1995) Multiple glucose 6-phosphate dehydrogenase–deficient variants correlate with malaria endemicity in the Vanuatu archipelago (southwestern Pacific). Am J Hum Genet 56:294–301 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Garcia A, Cot M, Chippaux JP, Ranque S, Feingold J, Demenais F, Abel L (1998 a ) Genetic control of blood infection levels in human malaria: evidence for a complex genetic model. Am J Trop Med Hyg 58:480–488 [ DOI ] [ PubMed ] [ Google Scholar ]
- Garcia A, Marquet S, Bucheton B, Hillaire D, Cot M, Fievet N, Dessein AJ, Abel L (1998 b ) Linkage analysis of blood Plasmodium falciparum levels: interest of the 5q31-q33 chromosome region. Am J Trop Med Hyg 58:705–709 [ DOI ] [ PubMed ] [ Google Scholar ]
- Genton B, al-Yaman F, Mgone CS, Alexander N, Paniu MM, Alpers MP, Mokela D (1995) Ovalocytosis and cerebral malaria. Nature 378:564–565 10.1038/378564a0 [ DOI ] [ PubMed ] [ Google Scholar ]
- Gilbert SC, Plebanski M, Gupta S, Morris J, Cox M, Aidoo M, Kwiatkowski D, Greenwood BM, Whittle HC, Hill AV (1998) Association of malaria parasite population structure, HLA, and immunological antagonism. Science 279:1173–1177 10.1126/science.279.5354.1173 [ DOI ] [ PubMed ] [ Google Scholar ]
- Gilles HM, Fletcher KA, Hendrickse RG, Lindner R, Reddy S, Allan N (1967) Glucose-6-phosphate-dehydrogenase deficiency, sickling, and malaria in African children in South Western Nigeria. Lancet 1:138–140 10.1016/S0140-6736(67)91037-9 [ DOI ] [ PubMed ] [ Google Scholar ]
- Grau GE, Fajardo LF, Piguet PF, Allet B, Lambert PH, Vassalli P (1987 a ) Tumor necrosis factor (cachectin) as an essential mediator in murine cerebral malaria. Science 237:1210–1212 [ DOI ] [ PubMed ] [ Google Scholar ]
- Grau GE, Gretener D, Lambert PH (1987 b ) Prevention of murine cerebral malaria by low-dose cyclosporin A. Immunology 61:521–525 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Grau GE, Heremans H, Piguet PF, Pointaire P, Lambert PH, Billiau A, Vassalli P (1989) Monoclonal antibody against interferon gamma can prevent experimental cerebral malaria and its associated overproduction of tumor necrosis factor. Proc Natl Acad Sci USA 86:5572–5574 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Greenberg J, Kendrick LP (1959) Resistance to malaria in hybrids between Swiss and certain other strains of mice. J Parasitol 45:263–267 [ PubMed ] [ Google Scholar ]
- Greenberg J, Nadel EM, Coatney GR (1954) Differences in survival of several inbred strains of mice and their hybrids infected with Plasmodium berghei. J Infect Dis 95:114–116 [ DOI ] [ PubMed ] [ Google Scholar ]
- Hadley TJ, Klotz FW, Pasvol G, Haynes JD, McGinniss MH, Okubo Y, Miller LH (1987) Falciparum malaria parasites invade erythrocytes that lack glycophorin A and B (MkMk): strain differences indicate receptor heterogeneity and two pathways for invasion. J Clin Invest 80:1190–1193 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Haldane JBS (1949) Disease and evolution. Ricerca Sci Suppl 19:3–10 [ Google Scholar ]
- Hamblin MT, Di Rienzo A (2000) Detection of the signature of natural selection in humans: evidence from the Duffy blood group locus. Am J Hum Genet 66:1669–1679 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Hamblin MT, Thompson EE, Di Rienzo A (2002) Complex signatures of natural selection at the Duffy blood group locus. Am J Hum Genet 70:369–383 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Hartl DL (2004) The origin of malaria: mixed messages from genetic diversity. Nat Rev Microbiol 2:15–22 10.1038/nrmicro795 [ DOI ] [ PubMed ] [ Google Scholar ]
- Hedrick P (2004) Estimation of relative fitnesses from relative risk data and the predicted future of haemoglobin alleles S and C. J Evol Biol 17:221–224 10.1046/j.1420-9101.2003.00635.x [ DOI ] [ PubMed ] [ Google Scholar ]
- Hernandez-Valladares M, Naessens J, Gibson JP, Musoke AJ, Nagda S, Rihet P, ole-MoiYoi OK, Iraqi FA (2004 a ) Confirmation and dissection of QTL controlling resistance to malaria in mice. Mamm Genome 15:390–398 10.1007/s00335-004-3042-4 [ DOI ] [ PubMed ] [ Google Scholar ]
- Hernandez-Valladares M, Rihet P, ole-MoiYoi OK, Iraqi FA (2004 b ) Mapping of a new quantitative trait locus for resistance to malaria in mice by a comparative mapping approach with human chromosome 5q31-q33. Immunogenetics 56:115–117 10.1007/s00251-004-0667-0 [ DOI ] [ PubMed ] [ Google Scholar ]
- Hill AV, Allsopp CE, Kwiatkowski D, Anstey NM, Twumasi P, Rowe PA, Bennett S, Brewster D, McMichael AJ, Greenwood BM (1991) Common West African HLA antigens are associated with protection from severe malaria. Nature 352:595–600 10.1038/352595a0 [ DOI ] [ PubMed ] [ Google Scholar ]
- Hill AVS, Elvin J, Willis AC, Aidoo M, Allsopp CEM, Gotch FM, Gao XM, Takiguchi M, Greenwood BM, Townsend ARM, McMichael AJ, Whittle HC (1992) Molecular analysis of the association of HLA-B53 and resistance to severe malaria. Nature 360:434–439 10.1038/360434a0 [ DOI ] [ PubMed ] [ Google Scholar ]
- Hobbs MR, Udhayakumar V, Levesque MC, Booth J, Roberts JM, Tkachuk AN, Pole A, Coon H, Kariuki S, Nahlen BL, Mwaikambo ED, Lal AL, Granger DL, Anstey NM, Weinberg JB (2002) A new NOS2 promoter polymorphism associated with increased nitric oxide production and protection from severe malaria in Tanzanian and Kenyan children. Lancet 360:1468–1475 10.1016/S0140-6736(02)11474-7 [ DOI ] [ PubMed ] [ Google Scholar ]
- Hunt NH, Driussi C, Sai-Kiang L (2001) Haptoglobin and malaria. Redox Rep 6:389–392 10.1179/135100001101536508 [ DOI ] [ PubMed ] [ Google Scholar ]
- Hunt NH, Manduci N, Thumwood CM (1993) Amelioration of murine cerebral malaria by dietary restriction. Parasitology 107:471–476 [ DOI ] [ PubMed ] [ Google Scholar ]
- Imrie H, Ferguson DJ, Day KP (2004) Human serum haptoglobin is toxic to Plasmodium falciparum in vitro. Mol Biochem Parasitol 133:93–98 10.1016/j.molbiopara.2003.07.007 [ DOI ] [ PubMed ] [ Google Scholar ]
- Jepson A, Sisay-Joof F, Banya W, Hassan-King M, Frodsham A, Bennett S, Hill AV, Whittle H (1997) Genetic linkage of mild malaria to the major histocompatibility complex in Gambian children: study of affected sibling pairs. BMJ 315:96–97 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Jepson AP, Banya WA, Sisay-Joof F, Hassan-King M, Bennett S, Whittle HC (1995) Genetic regulation of fever in Plasmodium falciparum malaria in Gambian twin children. J Infect Dis 172:316–319 [ DOI ] [ PubMed ] [ Google Scholar ]
- Joy DA, Feng X, Mu J, Furuya T, Chotivanich K, Krettli AU, Ho M, Wang A, White NJ, Suh E, Beerli P, Su XZ (2003) Early origin and recent expansion of Plasmodium falciparum. Science 300:318–321 10.1126/science.1081449 [ DOI ] [ PubMed ] [ Google Scholar ]
- Juliger S, Bongartz M, Luty AJ, Kremsner PG, Kun JF (2003) Functional analysis of a promoter variant of the gene encoding the interferon-gamma receptor chain I. Immunogenetics 54:675–680 [ DOI ] [ PubMed ] [ Google Scholar ]
- Kikuchi M, Looareesuwan S, Ubalee R, Tasanor O, Suzuki F, Wattanagoon Y, Na-Bangchang K, Kimura A, Aikawa M, Hirayama K (2001) Association of adhesion molecule PECAM-1/CD31 polymorphism with susceptibility to cerebral malaria in Thais. Parasitol Int 50:235–239 10.1016/S1383-5769(01)00082-4 [ DOI ] [ PubMed ] [ Google Scholar ]
- Knight JC, Keating BJ, Rockett KA, Kwiatkowski DP (2003) In vivo characterization of regulatory polymorphisms by allele-specific quantification of RNA polymerase loading. Nat Genet 33:469–475 10.1038/ng1124 [ DOI ] [ PubMed ] [ Google Scholar ]
- Knight JC, Udalova I, Hill AV, Greenwood BM, Peshu N, Marsh K, Kwiatkowski D (1999) A polymorphism that affects OCT-1 binding to the TNF promoter region is associated with severe malaria. Nat Genet 22:145–150 10.1038/9649 [ DOI ] [ PubMed ] [ Google Scholar ]
- Koch O, Awomoyi A, Usen S, Jallow M, Richardson A, Hull J, Pinder M, Newport M, Kwiatkowski D (2002) IFNGR1 gene promoter polymorphisms and susceptibility to cerebral malaria. J Infect Dis 185:1684–1687 [ DOI ] [ PubMed ] [ Google Scholar ]
- Koch O, Rockett K, Jallow M, Pinder M, Sisay-Joof F, Kwiatkowski D (2005) Investigation of malaria susceptibility determinants in the IFNG/IL26/IL22 genomic region. Genes Immun 6:312–318 10.1038/sj.gene.6364214 [ DOI ] [ PubMed ] [ Google Scholar ]
- Kremsner PG, Grundmann H, Neifer S, Sliwa K, Sahlmuller G, Hegenscheid B, Bienzle U (1991) Pentoxifylline prevents murine cerebral malaria. J Infect Dis 164:605–608 [ DOI ] [ PubMed ] [ Google Scholar ]
- Kun JF, Klabunde J, Lell B, Luckner D, Alpers M, May J, Meyer C, Kremsner PG (1999) Association of the ICAM-1 Kilifi mutation with protection against severe malaria in Lambarene, Gabon. Am J Trop Med Hyg 61:776–779 [ DOI ] [ PubMed ] [ Google Scholar ]
- Kun JF, Mordmuller B, Lell B, Lehman LG, Luckner D, Kremsner PG (1998) Polymorphism in promoter region of inducible nitric oxide synthase gene and protection against malaria. Lancet 351:265–266 10.1016/S0140-6736(05)78273-8 [ DOI ] [ PubMed ] [ Google Scholar ]
- Kun JF, Mordmuller B, Perkins DJ, May J, Mercereau-Puijalon O, Alpers M, Weinberg JB, Kremsner PG (2001) Nitric oxide synthase 2(Lambarene) (G-954C), increased nitric oxide production, and protection against malaria. J Infect Dis 184:330–336 [ DOI ] [ PubMed ] [ Google Scholar ]
- Kwiatkowski D (1995) Malarial toxins and the regulation of parasite density. Parasitol Today 11:206–212 10.1016/0169-4758(95)80079-4 [ DOI ] [ PubMed ] [ Google Scholar ]
- Kyes S, Horrocks P, Newbold C (2001) Antigenic variation at the infected red cell surface in malaria. Annu Rev Microbiol 55:673–707 10.1146/annurev.micro.55.1.673 [ DOI ] [ PubMed ] [ Google Scholar ]
- Lapoumeroulie C, Dunda O, Ducrocq R, Trabuchet G, Mony-Lobe M, Bodo JM, Carnevale P, Labie D, Elion J, Krishnamoorthy R (1992) A novel sickle cell mutation of yet another origin in Africa: the Cameroon type. Hum Genet 89:333–337 [ DOI ] [ PubMed ] [ Google Scholar ]
- Levesque MC, Hobbs MR, Anstey NM, Vaughn TN, Chancellor JA, Pole A, Perkins DJ, Misukonis MA, Chanock SJ, Granger DL, Weinberg JB (1999) Nitric oxide synthase type 2 promoter polymorphisms, nitric oxide production, and disease severity in Tanzanian children with malaria. J Infect Dis 180:1994–2002 [ DOI ] [ PubMed ] [ Google Scholar ]
- Luoni G, Verra F, Arca B, Sirima BS, Troye-Blomberg M, Coluzzi M, Kwiatkowski D, Modiano D (2001) Antimalarial antibody levels and IL4 polymorphism in the Fulani of West Africa. Genes Immun 2:411–414 10.1038/sj.gene.6363797 [ DOI ] [ PubMed ] [ Google Scholar ]
- Luty AJ, Kun JF, Kremsner PG (1998) Mannose-binding lectin plasma levels and gene polymorphisms in Plasmodium falciparum malaria. J Infect Dis 178:1221–1224 [ DOI ] [ PubMed ] [ Google Scholar ]
- Luzzatto L, Usanga FA, Reddy S (1969) Glucose-6-phosphate dehydrogenase deficient red cells: resistance to infection by malarial parasites. Science 164:839–842 [ DOI ] [ PubMed ] [ Google Scholar ]
- Mackinnon MJ, Gunawardena DM, Rajakaruna J, Weerasingha S, Mendis KN, Carter R (2000) Quantifying genetic and nongenetic contributions to malarial infection in a Sri Lankan population. Proc Natl Acad Sci USA 97:12661–12666 10.1073/pnas.220267997 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Maier AG, Duraisingh MT, Reeder JC, Patel SS, Kazura JW, Zimmerman PA, Cowman AF (2003) Plasmodium falciparum erythrocyte invasion through glycophorin C and selection for Gerbich negativity in human populations. Nat Med 9:87–92 10.1038/nm807 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Marsh K, Forster D, Waruiru C, Mwangi I, Winstanley M, Marsh V, Newton C, Winstanley P, Warn P, Peshu N, Pasvol G, Snow R (1995) Indicators of life-threatening malaria in African children. N Engl J Med 332:1399–1404 10.1056/NEJM199505253322102 [ DOI ] [ PubMed ] [ Google Scholar ]
- Mayor A, Bir N, Sawhney R, Singh S, Pattnaik P, Singh SK, Sharma A, Chitnis CE (2005) Receptor-binding residues lie in central regions of Duffy-binding-like domains involved in red cell invasion and cytoadherence by malaria parasites. Blood 105:2557–2563 10.1182/blood-2004-05-1722 [ DOI ] [ PubMed ] [ Google Scholar ]
- McGuire W, Hill AV, Allsopp CE, Greenwood BM, Kwiatkowski D (1994) Variation in the TNF-alpha promoter region associated with susceptibility to cerebral malaria. Nature 371:508–510 10.1038/371508a0 [ DOI ] [ PubMed ] [ Google Scholar ]
- McGuire W, Knight JC, Hill AV, Allsopp CE, Greenwood BM, Kwiatkowski D (1999) Severe malarial anemia and cerebral malaria are associated with different tumor necrosis factor promoter alleles. J Infect Dis 179:287–290 [ DOI ] [ PubMed ] [ Google Scholar ]
- Meyer CG, May J, Luty AJ, Lell B, Kremsner PG (2002) TNFalpha-308A associated with shorter intervals of Plasmodium falciparum reinfections. Tissue Antigens 59:287–292 10.1034/j.1399-0039.2002.590406.x [ DOI ] [ PubMed ] [ Google Scholar ]
- Michon P, Woolley I, Wood EM, Kastens W, Zimmerman PA, Adams JH (2001) Duffy-null promoter heterozygosity reduces DARC expression and abrogates adhesion of the P. vivax ligand required for blood-stage infection. FEBS Lett 495:111–114 10.1016/S0014-5793(01)02370-5 [ DOI ] [ PubMed ] [ Google Scholar ]
- Miller LH, Mason SJ, Clyde DF, McGinniss MH (1976) The resistance factor to Plasmodium vivax in blacks: the Duffy-blood-group genotype, FyFy. N Engl J Med 295:302–304 [ DOI ] [ PubMed ] [ Google Scholar ]
- Min-Oo G, Fortin A, Tam MF, Gros P, Stevenson MM (2004) Phenotypic expression of pyruvate kinase deficiency and protection against malaria in a mouse model. Genes Immun 5:168–175 10.1038/sj.gene.6364069 [ DOI ] [ PubMed ] [ Google Scholar ]
- Min-Oo G, Fortin A, Tam MF, Nantel A, Stevenson MM, Gros P (2003) Pyruvate kinase deficiency in mice protects against malaria. Nat Genet 35:357–362 10.1038/ng1260 [ DOI ] [ PubMed ] [ Google Scholar ]
- Mockenhaupt FP, Ehrhardt S, Cramer JP, Otchwemah RN, Anemana SD, Goltz K, Mylius F, Dietz E, Eggelte TA, Bienzle U (2004 a ) Hemoglobin C and resistance to severe malaria in Ghanaian children. J Infect Dis 190:1006–1009 [ DOI ] [ PubMed ] [ Google Scholar ]
- Mockenhaupt FP, Ehrhardt S, Gellert S, Otchwemah RN, Dietz E, Anemana SD, Bienzle U (2004 b ) Alpha( + )-thalassemia protects African children from severe malaria. Blood 104:2003–2006 10.1182/blood-2003-11-4090 [ DOI ] [ PubMed ] [ Google Scholar ]
- Modiano D, Chiucchiuini A, Petrarca V, Sirima BS, Luoni G, Perlmann H, Esposito F, Coluzzi M (1998) Humoral response to Plasmodium falciparum Pf155/ring-infected erythrocyte surface antigen and Pf332 in three sympatric ethnic groups of Burkina Faso. Am J Trop Med Hyg 58:220–224 [ DOI ] [ PubMed ] [ Google Scholar ]
- Modiano D, Chiucchiuini A, Petrarca V, Sirima BS, Luoni G, Roggero MA, Corradin G, Coluzzi M, Esposito F (1999) Interethnic differences in the humoral response to non-repetitive regions of the Plasmodium falciparum circumsporozoite protein. Am J Trop Med Hyg 61:663–667 [ DOI ] [ PubMed ] [ Google Scholar ]
- Modiano D, Luoni G, Sirima BS, Lanfrancotti A, Petrarca V, Cruciani F, Simpore J, Ciminelli BM, Foglietta E, Grisanti P, Bianco I, Modiano G, Coluzzi M (2001 a ) The lower susceptibility to Plasmodium falciparum malaria of Fulani of Burkina Faso (West Africa) is associated with low frequencies of classic malaria-resistance genes. Trans R Soc Trop Med Hyg 95:149–152 10.1016/S0035-9203(01)90141-5 [ DOI ] [ PubMed ] [ Google Scholar ]
- Modiano D, Luoni G, Sirima BS, Simpore J, Verra F, Konate A, Rastrelli E, Olivieri A, Calissano C, Paganotti GM, D’Urbano L, Sanou I, Sawadogo A, Modiano G, Coluzzi M (2001 b ) Haemoglobin C protects against clinical Plasmodium falciparum malaria. Nature 414:305–308 10.1038/35104556 [ DOI ] [ PubMed ] [ Google Scholar ]
- Modiano D, Petrarca V, Sirima BS, Nebie I, Diallo D, Esposito F, Coluzzi M (1996) Different response to Plasmodium falciparum malaria in West African sympatric ethnic groups. Proc Natl Acad Sci USA 93:13206–13211 10.1073/pnas.93.23.13206 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Modiano G, Morpurgo G, Terrenato L, Novelletto A, Di Rienzo A, Colombo B, Purpura M, Mariani M, Santachiara-Benerecetti S, Brega A, Dixit KA, Shrestha SL, Lania A, Wanachiwanawin W, Luzzatto L (1991) Protection against malaria morbidity: near-fixation of the α-thalassemia gene in a Nepalese population. Am J Hum Genet 48:390–397 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Morahan G, Boutlis CS, Huang D, Pain A, Saunders JR, Hobbs MR, Granger DL, Weinberg JB, Peshu N, Mwaikambo ED, Marsh K, Roberts DJ, Anstey NM (2002) A promoter polymorphism in the gene encoding interleukin-12 p40 (IL12B) is associated with mortality from cerebral malaria and with reduced nitric oxide production. Genes Immun 3:414–418 10.1038/sj.gene.6363909 [ DOI ] [ PubMed ] [ Google Scholar ]
- Nagayasu E, Ito M, Akaki M, Nakano Y, Kimura M, Looareesuwan S, Aikawa M (2001) CR1 density polymorphism on erythrocytes of falciparum malaria patients in Thailand. Am J Trop Med Hyg 64:1–5 [ DOI ] [ PubMed ] [ Google Scholar ]
- Nagayasu E, Nagakura K, Akaki M, Tamiya G, Makino S, Nakano Y, Kimura M, Aikawa M (2002) Association of a determinant on mouse chromosome 18 with experimental severe Plasmodium berghei malaria. Infect Immun 70:512–516 10.1128/IAI.70.2.512-516.2002 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Nagel RL, Ranney HM (1990) Genetic epidemiology of structural mutations of the beta-globin gene. Semin Hematol 27:342–359 [ PubMed ] [ Google Scholar ]
- Neill AL, Chan-Ling T, Hunt NH (1993) Comparisons between microvascular changes in cerebral and non-cerebral malaria in mice, using the retinal whole-mount technique. Parasitology 107:477–487 [ DOI ] [ PubMed ] [ Google Scholar ]
- Neill AL, Hunt NH (1992) Pathology of fatal and resolving Plasmodium berghei cerebral malaria in mice. Parasitology 105:165–175 [ DOI ] [ PubMed ] [ Google Scholar ]
- Ohashi J, Naka I, Patarapotikul J, Hananantachai H, Brittenham G, Looareesuwan S, Clark AG, Tokunaga K (2004) Extended linkage disequilibrium surrounding the hemoglobin E variant due to malarial selection. Am J Hum Genet 74:1198–1208 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Ohno T, Nishimura M (2004) Detection of a new cerebral malaria susceptibility locus, using CBA mice. Immunogenetics 56:675–678 10.1007/s00251-004-0739-1 [ DOI ] [ PubMed ] [ Google Scholar ]
- Omi K, Ohashi J, Patarapotikul J, Hananantachai H, Naka I, Looareesuwan S, Tokunaga K (2002) Fcgamma receptor IIA and IIIB polymorphisms are associated with susceptibility to cerebral malaria. Parasitol Int 51:361–366 10.1016/S1383-5769(02)00040-5 [ DOI ] [ PubMed ] [ Google Scholar ]
- ——— (2003) CD36 polymorphism is associated with protection from cerebral malaria. Am J Hum Genet 72:364–374 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Orlandi PA, Klotz FW, Haynes JD (1992) A malaria invasion receptor, the 175-kilodalton erythrocyte binding antigen of Plasmodium falciparum recognizes the terminal Neu5Ac(alpha 2–3)Gal- sequences of glycophorin A. J Cell Biol 116:901–909 10.1083/jcb.116.4.901 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Pain A, Urban BC, Kai O, Casals-Pascual C, Shafi J, Marsh K, Roberts DJ (2001) A non-sense mutation in Cd36 gene is associated with protection from severe malaria. Lancet 357:1502–1503 10.1016/S0140-6736(00)04662-6 [ DOI ] [ PubMed ] [ Google Scholar ]
- Pasvol G, Weatherall DJ, Wilson RJ (1978) Cellular mechanism for the protective effect of haemoglobin S against P. falciparum malaria. Nature 274:701–703 [ DOI ] [ PubMed ] [ Google Scholar ]
- Patel SS, King CL, Mgone CS, Kazura JW, Zimmerman PA (2004) Glycophorin C (Gerbich antigen blood group) and band 3 polymorphisms in two malaria holoendemic regions of Papua New Guinea. Am J Hematol 75:1–5 10.1002/ajh.10448 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Patel SS, Mehlotra RK, Kastens W, Mgone CS, Kazura JW, Zimmerman PA (2001) The association of the glycophorin C exon 3 deletion with ovalocytosis and malaria susceptibility in the Wosera, Papua New Guinea. Blood 98:3489–3491 10.1182/blood.V98.12.3489 [ DOI ] [ PubMed ] [ Google Scholar ]
- Pattanapanyasat K, Yongvanitchit K, Tongtawe P, Tachavanich K, Wanachiwanawin W, Fucharoen S, Walsh DS (1999) Impairment of Plasmodium falciparum growth in thalassemic red blood cells: further evidence by using biotin labeling and flow cytometry. Blood 93:3116–3119 [ PubMed ] [ Google Scholar ]
- Pauling L, Itano HA, Singer SJ, Wells IC (1949) Sickle cell anemia a molecular disease. Science 110:543–548 [ DOI ] [ PubMed ] [ Google Scholar ]
- Quaye IK, Ekuban FA, Goka BQ, Adabayeri V, Kurtzhals JA, Gyan B, Ankrah NA, Hviid L, Akanmori BD (2000) Haptoglobin 1-1 is associated with susceptibility to severe Plas modium falciparum malaria. Trans R Soc Trop Med Hyg 94:216–219 10.1016/S0035-9203(00)90281-5 [ DOI ] [ PubMed ] [ Google Scholar ]
- Ranque S, Safeukui I, Poudiougou B, Traore A, Keita M, Traore D, Diakite M, Cisse MB, Keita MM, Doumbo OK, Dessein AJ (2005) Familial aggregation of cerebral malaria and severe malarial anemia. J Infect Dis 191:799–804 [ DOI ] [ PubMed ] [ Google Scholar ]
- Rest JR (1982) Cerebral malaria in inbred mice. I. A new model and its pathology. Trans R Soc Trop Med Hyg 76:410–415 10.1016/0035-9203(82)90203-6 [ DOI ] [ PubMed ] [ Google Scholar ]
- Rihet P, Abel L, Traore Y, Traore-Leroux T, Aucan C, Fumoux F (1998 a ) Human malaria: segregation analysis of blood infection levels in a suburban area and a rural area in Burkina Faso. Genet Epidemiol 15:435–450 [ DOI ] [ PubMed ] [ Google Scholar ]
- Rihet P, Flori L, Tall F, Traore AS, Fumoux F (2004) Hemoglobin C is associated with reduced Plasmodium falciparum parasitemia and low risk of mild malaria attack. Hum Mol Genet 13:1–6 10.1093/hmg/ddh002 [ DOI ] [ PubMed ] [ Google Scholar ]
- Rihet P, Traore Y, Abel L, Aucan C, Traoré-Leroux T, Fumoux F (1998 b ) Malaria in humans: Plasmodium falciparum blood infection levels are linked to chromosome 5q31-q33. Am J Hum Genet 63:498–505 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Risch NJ (2000) Searching for genetic determinants in the new millennium. Nature 405:847–856 10.1038/35015718 [ DOI ] [ PubMed ] [ Google Scholar ]
- Roberts DJ, Craig AG, Berendt AR, Pinches R, Nash G, Marsh K, Newbold CI (1992) Rapid switching to multiple antigenic and adhesive phenotypes in malaria. Nature 357:689–692 10.1038/357689a0 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Rowe JA, Moulds JM, Newbold CI, Miller LH (1997) P. falciparum rosetting mediated by a parasite-variant erythrocyte membrane protein and complement-receptor 1. Nature 388:292–295 10.1038/40888 [ DOI ] [ PubMed ] [ Google Scholar ]
- Ruwende C, Hill A (1998) Glucose-6-phosphate dehydrogenase deficiency and malaria. J Mol Med 76:581–588 10.1007/s001090050253 [ DOI ] [ PubMed ] [ Google Scholar ]
- Ruwende C, Khoo SC, Snow RW, Yates SN, Kwiatkowski D, Gupta S, Warn P, Allsopp CE, Gilbert SC, Peschu N, Newbold CI, Greenwood BM, Marsh K, Hill AVS (1995) Natural selection of hemi- and heterozygotes for G6PD deficiency in Africa by resistance to severe malaria. Nature 376:246–249 10.1038/376246a0 [ DOI ] [ PubMed ] [ Google Scholar ]
- Sabeti P, Usen S, Farhadian S, Jallow M, Doherty T, Newport M, Pinder M, Ward R, Kwiatkowski D (2002 a ) CD40L association with protection from severe malaria. Genes Immun 3:286–291 10.1038/sj.gene.6363877 [ DOI ] [ PubMed ] [ Google Scholar ]
- Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, Ackerman HC, Campbell SJ, Altshuler D, Cooper R, Kwiatkowski D, Ward R, Lander ES (2002 b ) Detecting recent positive selection in the human genome from haplotype structure. Nature 419:832–837 10.1038/nature01140 [ DOI ] [ PubMed ] [ Google Scholar ]
- Sayles PC, Wassom DL (1988) Immunoregulation in murine malaria: susceptibility of inbred mice to infection with Plas modium yoelii depends on the dynamic interplay of host and parasite genes. J Immunol 141:241–248 [ PubMed ] [ Google Scholar ]
- Schofield L, Hewitt MC, Evans K, Siomos MA, Seeberger PH (2002) Synthetic GPI as a candidate anti-toxic vaccine in a model of malaria. Nature 418:785–789 10.1038/nature00937 [ DOI ] [ PubMed ] [ Google Scholar ]
- Sengupta S, Farheen S, Mukherjee N, Dey B, Mukhopadhyay B, Sil SK, Prabhakaran N, Ramesh A, Edwin D, Usha Rani MV, Mitra M, Mahadik CT, Singh S, Sehgal SC, Majumder PP (2004) DNA sequence variation and haplotype structure of the ICAM1 and TNF genes in 12 ethnic groups of India reveal patterns of importance in designing association studies. Ann Hum Genet 68:574–587 10.1046/j.1529-8817.2003.00126.x [ DOI ] [ PubMed ] [ Google Scholar ]
- Shear HL, Roth EF Jr, Fabry ME, Costantini FD, Pachnis A, Hood A, Nagel RL (1993) Transgenic mice expressing human sickle hemoglobin are partially resistant to rodent malaria. Blood 81:222–226 [ PubMed ] [ Google Scholar ]
- Shi YP, Nahlen BL, Kariuki S, Urdahl KB, McElroy PD, Roberts JM, Lal AA (2001) Fcgamma receptor IIa (CD32) polymorphism is associated with protection of infants against high-density Plasmodium falciparum infection. VII. Asembo Bay Cohort Project. J Infect Dis 184:107–111 [ DOI ] [ PubMed ] [ Google Scholar ]
- Sibley LD (2004) Intracellular parasite invasion strategies. Science 304:248–253 10.1126/science.1094717 [ DOI ] [ PubMed ] [ Google Scholar ]
- Sjoberg K, Lepers JP, Raharimalala L, Larsson A, Olerup O, Marbiah NT, Troye-Blomberg M, Perlmann P (1992) Genetic regulation of human anti-malarial antibodies in twins. Proc Natl Acad Sci USA 89:2101–2104 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI (2005) The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature 434:214–217 10.1038/nature03342 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Snow RW, Omumbo JA, Lowe B, Molyneux CS, Obiero JO, Palmer A, Weber MW, Pinder M, Nahlen B, Obonyo C, Newbold C, Gupta S, Marsh K (1997) Relation between severe malaria morbidity in children and level of Plasmodium falciparum transmission in Africa. Lancet 349:1650–1654 10.1016/S0140-6736(97)02038-2 [ DOI ] [ PubMed ] [ Google Scholar ]
- Sokhna CS, Rogier C, Dieye A, Trape JF (2000) Host factors affecting the delay of reappearance of Plasmodium falciparum after radical treatment among a semi-immune population exposed to intense perennial transmission. Am J Trop Med Hyg 62:266–270 [ DOI ] [ PubMed ] [ Google Scholar ]
- Stevenson MM, Lyanga JJ, Skamene E (1982) Murine malaria: genetic control of resistance to Plasmodium chabaudi. Infect Immun 38:80–88 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Stevenson MM, Riley EM (2004) Innate immunity to malaria. Nat Rev Immunol 4:169–180 10.1038/nri1311 [ DOI ] [ PubMed ] [ Google Scholar ]
- Stevenson MM, Skamene E (1985) Murine malaria: resistance of AXB/BXA recombinant inbred mice to Plasmodium chabaudi. Infect Immun 47:452–456 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Stirnadel HA, Al-Yaman F, Genton B, Alpers MP, Smith TA (2000 a ) Assessment of different sources of variation in the antibody responses to specific malaria antigens in children in Papua New Guinea. Int J Epidemiol 29:579–586 10.1093/ije/29.3.579 [ DOI ] [ PubMed ] [ Google Scholar ]
- Stirnadel HA, Beck HP, Alpers MP, Smith TA (1999 a ) Heritability and segregation analysis of immune responses to specific malaria antigens in Papua New Guinea. Genet Epidemiol 17:16–34 [ DOI ] [ PubMed ] [ Google Scholar ]
- ——— (2000 b ) Genetic analysis of IgG subclass responses against RESA and MSP2 of Plasmodium falciparum in adults in Papua New Guinea. Epidemiol Infect 124:153–162 10.1017/S0950268899003325 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Stirnadel HA, Stockle M, Felger I, Smith T, Tanner M, Beck HP (1999 b ) Malaria infection and morbidity in infants in relation to genetic polymorphisms in Tanzania. Trop Med Int Health 4:187–193 10.1046/j.1365-3156.1999.43381.x [ DOI ] [ PubMed ] [ Google Scholar ]
- Taylor TE, Fu WJ, Carr RA, Whitten RO, Mueller JS, Fosiko NG, Lewallen S, Liomba NG, Molyneux ME (2004) Differentiating the pathologies of cerebral malaria by postmortem parasite counts. Nat Med 10:143–145 10.1038/nm986 [ DOI ] [ PubMed ] [ Google Scholar ]
- Terrenato L, Shrestha S, Dixit KA, Luzzatto L, Modiano G, Morpurgo G, Arese P (1988) Decreased malaria morbidity in the Tharu people compared to sympatric populations in Nepal. Ann Trop Med Parasitol 82:1–11 [ DOI ] [ PubMed ] [ Google Scholar ]
- Thumwood CM, Hunt NH, Clark IA, Cowden WB (1988) Breakdown of the blood-brain barrier in murine cerebral malaria. Parasitology 96:579–589 [ DOI ] [ PubMed ] [ Google Scholar ]
- Tishkoff SA, Varkonyi R, Cahinhinan N, Abbes S, Argyropoulos G, Destro-Bisol G, Drousiotou A, Dangerfield B, Lefranc G, Loiselet J, Piro A, Stoneking M, Tagarelli A, Tagarelli G, Touma EH, Williams SM, Clark AG (2001) Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science 293:455–462 10.1126/science.1061573 [ DOI ] [ PubMed ] [ Google Scholar ]
- Tishkoff SA, Williams SM (2002) Genetic analysis of African populations: human evolution and complex disease. Nat Rev Genet 3:611–621 [ DOI ] [ PubMed ] [ Google Scholar ]
- Tokumasu F, Fairhurst RM, Ostera GR, Brittain NJ, Hwang J, Wellems TE, Dvorak JA (2005) Band 3 modifications in Plasmodium falciparum -infected AA and CC erythrocytes assayed by autocorrelation analysis using quantum dots. J Cell Sci 118:1091–1098 10.1242/jcs.01662 [ DOI ] [ PubMed ] [ Google Scholar ]
- Tournamille C, Colin Y, Cartron JP, Le Van Kim C (1995) Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet 10:224–228 [ DOI ] [ PubMed ] [ Google Scholar ]
- Trape JF, Rogier C, Konate L, Diagne N, Bouganali H, Canque B, Legros F, Badji A, Ndiaye G, Ndiaye P, Brahimi K, Faye O, Druilhe P, Pereira da Silva L (1994) The Dielmo project: a longitudinal study of natural malaria infection and the mechanisms of protective immunity in a community living in a holoendemic area of Senegal. Am J Trop Med Hyg 51:123–137 [ DOI ] [ PubMed ] [ Google Scholar ]
- Treutiger CJ, Heddini A, Fernandez V, Muller WA, Wahlgren M (1997) PECAM-1/CD31, an endothelial receptor for binding Plasmodium falciparum -infected erythrocytes. Nat Med 3:1405–1408 10.1038/nm1297-1405 [ DOI ] [ PubMed ] [ Google Scholar ]
- Urban BC, Ferguson DJ, Pain A, Willcox N, Plebanski M, Austyn JM, Roberts DJ (1999) Plasmodium falciparum -infected erythrocytes modulate the maturation of dendritic cells. Nature 400:73–77 10.1038/21900 [ DOI ] [ PubMed ] [ Google Scholar ]
- Urban BC, Willcox N, Roberts DJ (2001) A role for CD36 in the regulation of dendritic cell function. Proc Natl Acad Sci USA 98:8750–8755 10.1073/pnas.151028698 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Usanga EA, Luzzatto L (1985) Adaptation of Plasmodium fal ciparum to glucose 6-phosphate dehydrogenase-deficient host red cells by production of parasite-encoded enzyme. Nature 313:793–795 10.1038/313793a0 [ DOI ] [ PubMed ] [ Google Scholar ]
- Vigario AM, Belnoue E, Cumano A, Marussig M, Miltgen F, Landau I, Mazier D, Gresser I, Renia L (2001) Inhibition of Plasmodium yoelii blood-stage malaria by interferon alpha through the inhibition of the production of its target cell, the reticulocyte. Blood 97:3966–3971 10.1182/blood.V97.12.3966 [ DOI ] [ PubMed ] [ Google Scholar ]
- Walley AJ, Aucan C, Kwiatkowski D, Hill AV (2004) Interleukin-1 gene cluster polymorphisms and susceptibility to clinical malaria in a Gambian case-control study. Eur J Hum Genet 12:132–138 10.1038/sj.ejhg.5201084 [ DOI ] [ PubMed ] [ Google Scholar ]
- Wang HY, Tang H, Shen CK, Wu CI (2003) Rapidly evolving genes in human. I. The glycophorins and their possible role in evading malaria parasites. Mol Biol Evol 20:1795–1804 10.1093/molbev/msg185 [ DOI ] [ PubMed ] [ Google Scholar ]
- Wattavidanage J, Carter R, Perera KL, Munasingha A, Bandara S, McGuinness D, Wickramasinghe AR, Alles HK, Mendis KN, Premawansa S (1999) TNFalpha*2 marks high risk of severe disease during Plasmodium falciparum malaria and other infections in Sri Lankans. Clin Exp Immunol 115:350–355 10.1046/j.1365-2249.1999.00804.x [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Weatherall DJ, Clegg JB (2001) Inherited haemoglobin disorders: an increasing global health problem. Bull World Health Organ 79:704–712 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Westendorp RG, van Dunne FM, Kirkwood TB, Helmerhorst FM, Huizinga TW (2001) Optimizing human fertility and survival. Nat Med 7:873 10.1038/90868 [ DOI ] [ PubMed ] [ Google Scholar ]
- Williams TN, Maitland K, Bennett S, Ganczakowski M, Peto TE, Newbold CI, Bowden DK, Weatherall DJ, Clegg JB (1996) High incidence of malaria in alpha-thalassaemic children. Nature 383:522–525 10.1038/383522a0 [ DOI ] [ PubMed ] [ Google Scholar ]
- Williams TN, Mwangi TW, Roberts DJ, Alexander ND, Weatherall DJ, Wambua S, Kortok M, Snow RW, Marsh K (2005 a ) An immune basis for malaria protection by the sickle cell trait. PLoS Med 2:e128 10.1371/journal.pmed.0020128 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Williams TN, Wambua S, Uyoga S, Macharia A, Mwacharo JK, Newton CR, Maitland K (2005 b ) Both heterozygous and homozygous α + thalassemia protect against severe and fatal Plasmodium falciparum malaria on the coast of Kenya. Blood 106:368–371 10.1182/blood-2005-01-0313 [ DOI ] [ PubMed ] [ Google Scholar ]
- Williams TN, Weatherall DJ, Newbold CI (2002) The membrane characteristics of Plasmodium falciparum -infected and -uninfected heterozygous alpha (0) thalassaemic erythrocytes. Br J Haematol 118:663–670 10.1046/j.1365-2141.2002.03610.x [ DOI ] [ PubMed ] [ Google Scholar ]
- Wilson J, Rockett K, Jallow M, Pinder M, Sisay-Joof F, Newport M, Newton J, Kwiatkowski D (2005) Analysis of IL10 haplotypic associations with severe malaria. Genes Immun ( http://www.nature.com/gene/journal/vaop/ncurrent/full/6364227a.html ) (electronically published June 3, 2005; accessed June 16, 2005) [ DOI ] [ PubMed ] [ Google Scholar ]
- Yazdani SS, Shakri AR, Mukherjee P, Baniwal SK, Chitnis CE (2004) Evaluation of immune responses elicited in mice against a recombinant malaria vaccine based on Plasmodium vivax Duffy binding protein. Vaccine 22:3727–3737 10.1016/j.vaccine.2004.03.030 [ DOI ] [ PubMed ] [ Google Scholar ]
- Zimmerman PA, Fitness J, Moulds JM, McNamara DT, Kasehagen LJ, Rowe JA, Hill AV (2003) CR1 Knops blood group alleles are not associated with severe malaria in the Gambia. Genes Immun 4:368–373 10.1038/sj.gene.6363980 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Zimmerman PA, Woolley I, Masinde GL, Miller SM, McNamara DT, Hazlett F, Mgone CS, Alpers MP, Genton B, Boatin BA, Kazura JW (1999) Emergence of FY*A(null) in a Plasmodium vivax -endemic region of Papua New Guinea. Proc Natl Acad Sci USA 96:13973–13977 10.1073/pnas.96.24.13973 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- View on publisher site
- PDF (188.8 KB)
- Collections
Similar articles
Cited by other articles, links to ncbi databases.
- Download .nbib .nbib
- Format: AMA APA MLA NLM
Add to Collections
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Published: 23 March 2011
Population genetics of malaria resistance in humans
- P W Hedrick 1
Heredity volume 107 , pages 283–304 ( 2011 ) Cite this article
18k Accesses
190 Citations
13 Altmetric
Metrics details
- Haematological diseases
- Population genetics
A Corrigendum to this article was published on 16 November 2011
The high mortality and widespread impact of malaria have resulted in this disease being the strongest evolutionary selective force in recent human history, and genes that confer resistance to malaria provide some of the best-known case studies of strong positive selection in modern humans. I begin by reviewing JBS Haldane's initial contribution to the potential of malaria genetic resistance in humans. Further, I discuss the population genetics aspects of many of the variants, including globin, G6PD deficiency, Duffy, ovalocytosis, ABO and human leukocyte antigen variants. Many of the variants conferring resistance to malaria are ‘loss-of-function’ mutants and appear to be recent polymorphisms from the last 5000–10 000 years or less. I discuss estimation of selection coefficients from case–control data and make predictions about the change for S , C and G6PD-deficiency variants. In addition, I consider the predicted joint changes when the two β-globin alleles S and C are both variable in the same population and when there is a variation for α-thalassemia and S , two unlinked, but epistatic variants. As more becomes known about genes conferring genetic resistance to malaria in humans, population genetics approaches can contribute both to investigating past selection and predicting the consequences in future generations for these variants.
Similar content being viewed by others

Malaria protection due to sickle haemoglobin depends on parasite genotype
The effect on the equilibrium sickle cell allele frequency of the probable protection conferred by malaria and sickle cell gene against other infectious diseases
On the feasibility of malaria hypothesis
Introduction.
Today, the knowledge that specific genetic variants can contribute to disease resistance in humans is well known ( Dronamraju, 2004 ; Kaslow et al., 2008 ). Haldane (1949a) is widely credited with first proposing that genetic resistance to a disease was potentially an important evolutionary force in humans. Before discussing malaria resistance further, it is worthwhile to consider the initial historical contribution and insights of JBS Haldane, one of the founders of population genetics, to the population genetics of malaria resistance in humans.
Acknowledgment to the general review entitled ‘Disease and Evolution’ by Haldane (1949a) is confusing because there is nothing discussed about human resistance to disease in the body of this manuscript from a meeting in 1948 in Milan. Only in the subsequent discussion from G Montalenti at the end of the article (in Italian), and then from Haldane's response to Montalenti's comment, is this hypothesis put forward. As a result, it is useful to present below a translation of these comments from the Italian and Haldane's brief response.
MONTALENTI. Stresses the importance of the views expressed by Professor Haldane. Recalls the case of microcitemia or thalassemia studied by Silvestroni, Bianco and Montalenti. Recalls a gene, lethal in the homozygous state (Cooley's disease) occurs, in the heterozygous state, with such frequency in some populations (more than 10%) that one must admit that it represents in this condition an advantage for individuals that carry it. Because of some research, far from complete, it appears that the gene is more frequent in malarial areas. Professor Haldane has suggested in verbal communication that individuals with microcitemico, who among other characteristics have increased globular resistance, may be more resistant to malaria. However, this is an interesting case of heterosis, which relates to what was described by Professor Haldane.
HALDANE. 1: I agree with Dr Montalenti's project. Another possibility is that (by analogy with the advantage possessed by vermillion Drosophila on media deficient in tryptophan) microcythemic heterozygotes may be at an advantage on diets deficient in iron or other substances, thus leading to anaemia.
In addition, at the VIII International Congress of Genetics in Stockholm in 1948, actually previous to the Milan meeting, Haldane (1949b) outlined his hypothesis in more detail. This was in response to the idea put forth by Neel and Valentine (1947) that different human populations may have different mutation rates.
Finally, Neel and Valentine (1947) have calculated a mutation rate for the gene which when homozygous gives rise to the lethal condition of thalassemia major (Cooley's anemia), while the heterozygote has a mild microcytic anemia with decreased fragility of the erythrocytes. The gene is found in about 4.1% of the people of Italian origin in Rochester, NY. If the heterozygote had normal viability, equilibrium would be secured by a mutation rate of 4 × 10 −4 . On the other hand, if the heterozygotes had an increased fitness of only 2.1% there would be equilibrium without any mutation at all.
Neel and Valentine believe that the heterozygote is less fit than normal, and think that the mutation rate is above 4 × 10 −4 rather than below it. I believe that the possibility that the heterozygote is fitter than normal must be seriously considered. Such increased fitness is found in the case of several lethal and sublethal genes in Drosophila and Zea . A possible mechanism is as follows. The corpuscles of the anemic heterozygotes are smaller than normal and more resistant to hypotonic solutions. It is at least conceivable that they are also more resistant to attacks by the sporozoa that cause malaria, a disease prevalent in Italy, Sicily and Greece, where the gene is frequent. Similarly, the gene, which causes an anemia similar to that of iron deficiency, might be harmless or even useful to persons on an iron-deficient diet, although causing a relative anemia when the diet is more generous. Numerous other similar hypotheses could be framed. On many of them, the gene would lower fitness in America, but might, at least in the heterozygous condition, have the opposite effect in Greece or Sicily. Until more is known of the physiology of this gene in various environments, I doubt if we can accept the hypothesis that it arises very frequently by mutation in a small section of the human species.
From examining these passages, it seems appropriate to cite Haldane (1949b) as the first source of the idea that genetic variants in humans may confer resistance to malaria (see also the discussion in Crow, 2004 ; Weatherall, 2004 ).
At present, it is difficult to imagine that Haldane in 1948 was the first to articulate the potential importance of genetic variants in humans for resistance to malaria (actually, Beet, 1946 found that there were lower rates of malaria in sickle-cell heterozygotes than in normal individuals). The first generally recognized evidence for genetic resistance to malaria was published only a few years later by Allison (1954 , 2004 ) showing that the densities of the malarial parasite, Plasmodium falciparum , were significantly lower in sickle-cell hemoglobin heterozygotes AS than in normal AA individuals. Allison (1955) then discussed the evolutionary implications of his findings in the influential 1955 Cold Springs Harbor Symposium on Quantitative Biology, probably the initiation of the large effort, both medically and evolutionarily, to understand genetic resistance to malaria in humans (see also Allison, 1964 ). Overall, the original ‘malaria hypothesis’ of Haldane that diseases such as thalassemia are polymorphisms with an advantage to heterozygotes because of the balance of the benefits of resistance to malaria and detrimental effects due to the disease has been proven correct; however, the actual mechanisms of protection for different genetic variants have often proven to be complicated to understand.
My purpose here is to discuss some of the recent evolutionary genetic developments in human genetic resistance to malaria. Fortunately, in recent years there have been excellent reviews of the mechanisms of malaria resistance, as well as surveys of the many genes that may putatively confer malaria resistance ( Kwiatkowski, 2005 ; Verra et al., 2009 ; López et al., 2010 ). Therefore, I will not discuss in detail the molecular basis of the mechanisms of resistance or enumerate all the potential malaria resistance genes, but will focus on the genetic variants in which there is strong evidence for resistance, and data useful for population genetic approaches and analysis. Understanding the geographic patterns of these variants is fundamentally influenced by resistance selection, but would also include consideration of gene flow, mutation, genetic drift and the interaction of all these factors. I will mention these other evolutionary factors as appropriate, but will focus here on the effect of selection on these advantageous variants. I will also discuss the population genetics of two examples in which there are two malarial resistance variants in the same population, the two alleles S and C at the β-globin locus and then alleles at two different genes, the S allele at the β-globin locus and the α + thalassemia variant at the α-globin locus.
General background
Surprising to many today, malaria was endemic in southern parts of Europe and North America less than a century ago and was not eliminated from many locations around the Mediterranean Sea until after World War II in the late 1940s. Today malaria is still found across most of sub-Saharan Africa and in extensive regions in Asia and South America. To illustrate the human impact of malaria, in 2008 there were 243 million clinical cases and an estimated 863 000 deaths from malaria ( WHO, 2009 ), many of whom were children, making it one of the leading causes of death worldwide. Most of this mortality is from ‘severe malaria,’ which is due to P. falciparum , begins about 6–14 days after infection from mosquitoes, has the biggest impact on children and pregnant women, progresses rapidly and can cause death within hours or days (here, I will generally be considering the impact of severe malaria). The past mortality rate from malaria was even higher, particularly for populations without previous exposure to malaria. For example, on the west coasts of Africa in the early 1800s, mortality rates in Europeans often exceeded 50% per year ( Curtin, 1989 ). After quinine was introduced in the later 1800s, the mortality rate reduced to about 25% of that before its introduction, indicating that the great majority of the mortality was due to malaria.
It is generally assumed that death from malaria increased between 10 000 and 5000 years ago with the development of agriculture and settlements. These changes are thought to have increased both the density of the malaria mosquito vector and human population density, thereby greatly facilitating the transmission of malaria ( Carter and Mendis, 2002 ). Support for this hypothesis comes from some molecular analysis of P. falciparum strains that suggests that the African malaria population expanded around 10 000 years ago and spread to other areas ( Joy et al., 2003 ). In addition, chromosomal data from the mosquito vector Anopheles gambiae support contemporaneous speciation ( Coluzzi et al., 2002 see also; Lawniczak et al., 2010 and Neafsey et al., 2010 ). Recent analysis suggests that P. falciparum in humans is the result of cross-species transmission from gorillas ( Liu et al., 2010 ). Although it is not possible to date this transmission precisely at this point, it appears recent and may be coincidental with the advent of strong selection for malaria resistance in humans.
Although I will generally not discuss the mosquito vector of malaria here, Carter and Mendis (2002) summarized some relevant information about vector–host preference. In most areas where endemic malaria is prevalent, Anopheles mosquitoes are generally highly zoophilic, rather than anthropophilic, that is, they greatly prefer animals to humans for blood meals. However, in sub-Saharan Africa, these mosquito vectors are highly anthropophilic. Livingstone (1958) suggested that this was an adaptation to preference for humans in the new agrarian environments that had both high densities of humans and provided numerous small water pools for mosquito development. On the other hand, in Asia and the Middle East, there was an abundance of domesticated animal species during the rise of agriculture, which served as a blood source for mosquitoes; there were generally no such domesticated animal sources in Africa.
P. falciparum is the malarial species that is the most important cause of mortality in humans but, in addition, nearly 3 billion people, mainly in Asia and South America, are at risk from another malaria species, P. vivax , which causes chronic disease and deadly complications ( Guerra et al., 2010 ). Three more species, P. ovale , P. malariae and P. knowlesi ( Cox-Singh et al., 2008 ) can also cause malaria in humans. Infections with malaria may reach very high levels, for example, Mueller et al. (2009) found that 73% of a Papua New Guinea population were infected. When the species of malaria infecting individuals were identified, 54.9, 35.7, 13.4 and 4.8% of the individuals were infected by P. falciparum , P. vivax , P.malariae and P. ovale , respectively, and a significant excess of mixed infections over expectations was detected.
As a result of the high mortality and widespread impact of malaria, it is thought to be the strongest evolutionary selective force in recent human history ( Kwiatkowski, 2005 ). In fact, genes that confer resistance to malaria provide some of the best-known case studies of strong positive selection in modern humans. For example, maintenance of the sickle-cell hemoglobin variant is the classic example of heterozygote advantage, G6PD deficiency is an example of very strong selection at an X-linked locus, variants at β-globin ( S , C and E ) and those causing G6PD deficiency ( A -, Med and Mahidol ) illustrate selective advantage from a single-nucleotide change, HLA-B53 provides an example of a selective advantage of a gene conversion product and α-thalassemia provides an example of selection on duplicated loci. Before I examine resistance from some specific genetic variants, let me summarize several important general findings related to genetic resistance to malaria in humans.
(1) There is extensive overlap of the historical geographical distribution of malaria and human genetic variants that confer resistance to malaria. For example, P. falciparum is found across Africa and Asia, as are the resistance variants of hemoglobin and G6PD. Further, in regions of high endemic malaria, such as sub-Saharan tropical Africa and lowland Melanesia, there are generally more than one resistant variant present. Resistant variants are generally not present in regions where there is no history of malaria, except in recent immigrants ( O'Shaughnessy et al., 1990 ). For example, there does not appear to be any of the common malaria-resistant variants in native people of the new world ( Livingstone, 1985 ), apparently because their Asian ancestors were not exposed to malaria and malaria was only brought to the new world by early Spanish explorers in the 1600s and 1700s. In addition, there are convincing examples of microgeographic variation in β-thalassemia with higher frequencies at lower altitudes in Sardinia where malaria was historically endemic compared with higher altitudes ( Siniscalco et al., 1961 ) and variation of α-thalassemia ( Flint et al., 1986 ) and β-thalassemia ( Hill et al., 1988 ) in Melanesia correlated with altitude, which is highly correlated with malaria endemicity.
(2) The presence of strong selective pressure for resistance to malaria has resulted in the high frequency of some detrimental genetic diseases, such as sickle-cell anemia, thalassemia, G6PD deficiency and ovalocytosis. In fact, Weatherall (2010) has suggested that there is now a global health burden because more than 300 000 children are born each year with a severe inherited hemoglobin disorder that is in high frequency because it confers, or has conferred, malarial resistance. In fact, this strong selective pressure explains why disorders of the red blood cell are by far the most common genetic diseases in humans ( Weatherall, 2008 ). Further, it is possible that some of our other immunological, inflammatory and other chronic diseases found today may be due to the pleiotropic effects of the malaria resistance variants in high frequency because of the evolutionary pressure that malaria has exerted on humans ( Kwiatkowski, 2005 ).
(3) Many of the variants conferring resistance to malaria are ‘loss-of-function’ mutants and result in changed, generally reduced, expression or an altered product. In some cases, these mutants have a large pleiotropic cost associated with them and result in disease, as discussed above. In other cases, the change in expression or in gene product does not appear to have a major cost, as in the Duffy ‘null’ variants, the O in the ABO system and allele C for the β-globin gene.
(4) Variants at some loci conferring resistance to malaria are nucleotide- or codon-specific whereas others are quite general. For example, the two known independent Duffy ‘null’ variants in African and Papua New Guinea are at the exact same nucleotide position, 33 nucleotides upstream from the start codon apparently on different genetic backgrounds. Moreover, two of the three structural polymorphic β-globin variants that confer resistance to malaria, S and C , are at the same codon. The only other structural variant that appears to provide resistance to malaria, E , may provide protection because it alters β-globin expression and functions similar to a thalassemia variant. On the other hand, there are multiple variants that alter expression levels and provide protection within each of the categories known as α-thalassemia, β-thalassemia and G6PD deficiency.
(5) It appears that many of the genetic variants that provide resistance to severe malaria that have been dated are recent, not ancient, polymorphisms and that they have arisen in the last 5000–10 000 years or less (see Table 2). That is, these variants appear to date to a recent time when severe malaria became a strong selective force in humans. This means that these variants did not provide resistance to malaria in other species, that is, they are not trans-specific polymorphisms, and that they have been generated by new mutations in our recent human ancestors. Either there does not appear to be homologous resistance variants in other primates ( Verrelli et al., 2006 ; MacFie et al., 2009 ) or the malaria resistance variants found in primates are not identical to those in humans ( Tung et al., 2009 ).
(6) There are many different unlinked genes that appear to confer resistance to malaria in humans. Because the selection pressure from malaria through high mortality and widespread exposure is great, genes that provide resistance to malaria would be selected even though there may be a cost to resistance from genetic disease. In addition, there appears to be many different types of genes that can confer resistance. For example, Kwiatkowski and Luoni (2008) list potential resistance genes in the categories of hemoglobin genes, erythrocyte surface molecules, other erythrocyte-related proteins, adhesion molecules, mediators of innate immunity and acquired immunity genes. Recent genome-wide studies show the potential to identify many other genes that have a smaller effect on malarial resistance. Further, although much of the variation in malaria resistance appears because of genetic effects, only a small proportion of this variation appears because of well-known genetic factors, such as sickle-cell and α-thalassemia ( Mackinnon et al., 2005 ).
(7) Some of the genes that confer resistance to malaria are among the most variable genes in the human genome. The human leukocyte antigen (HLA), G6PD, globin and ABO genes are all among the most variable genes in the human genome and all appear to have variants that confer resistance to malaria. This high variation may be because genomic surveys have shown that disease resistance genes in general have high standing variation and that some of this variability provides resistance to malaria. It may also be because these regions have a high enough mutation rate by which they can generate enough variants quickly enough to provide variation for positive selection for malaria resistance.
(8) Different populations appear to have different levels of resistance to malaria even when they both live in the same, or similar, area where malaria is prevalent. These differences could result from non-genetic factors such as culture, nutritional status or patterns of disease transmission or incidence, but in some cases they appear to be from genetic differences such as different frequencies of resistance variants (for example, high frequency of Duffy ‘null’ in Africa confers resistance to P. vivax , see below; high frequency of α-thalassemia in Nepalese populations, Modiano et al., 1991 ), epistasis with other host resistance variants or differences in unknown genetic resistance factors ( Dolo et al., 2005 ).
Resistance mutants
Many different genes are involved with genetic resistance to malaria ( Kwiatkowski, 2005 ; Verra et al., 2009 ) and these genetic factors may vary over populations because of the presence or absence of specific resistant genetic variants, different species or strains of malaria and different effects of specific variants in given host genotypes. Other factors such as population structure and chance, as well as the experimental design or the phenotype(s) of malaria diagnosed in a given study, may impact the conclusions. Efforts to use genomics approaches and genome-wide association analysis have been initiated ( Ayodo et al., 2007 ; Timmann et al., 2007 ; The Malaria Genomic Epidemiology Network, 2008 ; Jallow et al., 2009 ; Eid et al., 2010 ) and some new regions of potential malaria resistance have been identified.
In an estimate of the extent of malaria resistance explained by genetic factors in Kenya using pedigree analysis, 25% was attributed to additively functioning host genes and 29% to unidentified household effects ( Mackinnon et al., 2005 ). Only 2.1% of the variation was attributed to sickle-cell variation and, using an unrelated approach, only about 2% was attributed to α-thalassemia. In other words, a large part of the variation in malaria resistance was attributed to additive genetic effects, but only a small proportion could be attributed to the most well-known genetic factors in the population.
Below I will concentrate on genes for which there is strong evidence for resistance and data that are useful for population genetic approaches and analysis. In general, I will discuss resistance variants that lower mortality from severe malaria, although resistance variants may reduce mortality or morbidity when malaria is not severe or have other impacts, such as reduction in the rate of malaria infection. Some other important genes not discussed here are those that are concerned with the innate immune response (as distinct from the innate resistance) and cytoadherence ( Kwiatkowski and Luoni, 2008 ). Some recent references to genes that are not discussed here are for the tumor necrosis factor ( TNF ) gene ( Clark et al., 2009 ), the toll-like receptor ( TLR ) genes ( Ferwerda et al., 2007 ) and the genes that cause CD36 deficiency ( Fry et al., 2009 ).
Hemoglobin mutants
Hemoglobinopathies, inherited disorders of hemoglobin structure and function, consist of structural hemoglobin variants and the thalassemias, inherited defects that reduce production in the synthesis of the α- and β-globins of the human adult hemoglobin ( Weatherall and Clegg, 2001 ; Weatherall, 2008 ). Two α and two β subunits compose the tetrameric protein backbone of adult hemoglobin, and the α and β subunits are made up of 141 and 146 amino acids, respectively, each with three exons. The genes that code for α- and β-globins, HBA (there are actually two genes, HBA1 and HBA2 , with identical amino-acid sequences) and HBB , are unlinked and are located on chromosomes 11 and 16, respectively. Hundreds of structural variants of adult hemoglobin have been documented ( Giardine et al., 2007 ), but only three, all at the HBB locus, S , C and E , have reached substantial frequencies in any populations. Two of these three, S and C , are different mutants (with different phenotypes) at the same amino-acid position 6 and, although E is a structural variant at amino-acid position 26, it also affects regulation in a manner similar to the thalassemias. Surprisingly then, the apparently advantageous structural variants are only at two codons in gene HBB , suggesting that mutants at other sites at the HBB locus, and all sites at the HBA loci, are either neutral or detrimental in malarial environments (or have not undergone mutation in malarial environments). Finally, every region in the world where malaria was prevalent has hemoglobinopathies, but no two areas have the same hemoglobinopathy or combination of hemoglobinopathies ( Flint et al., 1998 ). I will discuss some of these patterns below, but here I suggest that they appear to be caused primarily by differential mutation originating in various regions where malaria is prevalent and potentially by subsequent selective interaction between the variants.
Sickle cell, S or β S or HbS
The sickle-cell allele (for simplicity I will symbolize it by S ) was one of the first human genetic variants that was associated with a specific molecular-caused disease ( Pauling et al., 1949 ; Ingram, 2004 ). The sickle-cell allele is widely known as a variant that causes red blood cells to be deformed into a sickle shape when deoxygenated in AS heterozygotes, in which A indicates the non-mutant form of the β-globin gene, and also provides resistance to malaria in AS heterozygotes. In SS homozygotes, S causes the severe disease sickle-cell anemia. It is generally assumed that individuals with genotype SS had very low or zero fitness when, or if, there was no modern medical care. The greatest documented impact of S is that it protects against death or severe malarial disease in heterozygotes, and it appears that S has less effect on the rate of infection ( Weatherall, 2008 ). Multiple mechanisms of malaria protection in AS individuals have been proposed and supported, including that the growth of malarial parasites is suppressed in sickle cells.
The S allele differs from the normal A allele at one nucleotide at codon position 6 in the β-globin molecule, which translates into a change from the amino acid glutamic acid to valine (β6 Glu → Val and GAG → GTG; Table 1 ). The frequency of allele S is up to 0.2 in some parts of sub-Saharan Africa, Greece and India. There appears to be five independent origins, or mutations, of the S allele from the A allele, based on their existence in nearly non-overlapping geographical regions, four in Africa, Bantu (Central Africa Republic, CAR), Benin, Cameroon and Senegal, and another one in both India and Saudi Arabia, and the extensive differences in linked variants resulting in association with different classical haplotypes. Mediterranean S alleles appear to descend from the African S alleles, many of them from the Benin haplotype ( Flint et al., 1998 ). Recently, Ralph and Coop (2010) have shown that such parallel adaptation of multiple S alleles is not inconsistent with their theoretical spatial model and general estimates of mutation rate, gene flow, selection and population size, a finding consistent with that of Karasov et al. (2010) who suggest that mutation does not limit adaptation in many situations. On the other hand, it has been proposed that a plausible explanation for much of the haplotype diversity around S alleles is that it reflects past recombination and gene conversion ( Flint et al., 1998 ).
A general approach to determine the risk of individuals with a given genotype getting a disease, relative to that in the rest of the population, is to calculate the odds ratio ( OR ) as
where f c and f d are the frequencies of the genotype in control and diseased groups, respectively. Another measure of the protectiveness of a genotype from a disease is the relative risk ( RR ), which is
OR and RR are closest to each other when both f c and f d are in low frequency (for an introduction to OR and RR , see Agresti, 2007 ).
In a large study of HLA variation and malaria in Gambia, Hill et al. (1991) also examined the effect of sickle-cell variation on the presence of severe malaria as a standard to measure the impact of HLA variation. In 619 children with severe malaria only seven (0.012) were AS , carriers of the S allele. On the other hand, in 510 other children, who were outpatients and termed ‘mild controls’, many more, that is, 66 (0.129) were AS . Further in this study, OR =0.082 and RR =0.093, indicating very great protection (often calculated as 1— OR or 1— RR ) from severe malaria for AS genotypes. Ackerman et al. (2005) compared this ‘case–control’ method with a family-based method in more recent samples from Gambia and found similar OR values of 0.10 and 0.11.
To understand the dynamics of genetic change from adaptation for resistance to malaria, it is useful to estimate the selection coefficients for the different genotypes, values that can be estimated from OR data; but first, let me show how OR can be used in a population genetics context. Let me define the probability that malaria is present, given the AS heterozygote, relative to the probability of malaria when another genotype is present, as w . Using the traditional population genetics equation for viability selection (without reproduction; Hedrick 2011a , p. 129), the frequency of the genotype AS in the disease group is expected to be
This expression can be solved for w and is identical to OR
The selection coefficient ( s ) for genotype AS can be estimated as
where m is the rate of non-genotype-specific mortality due to malaria (generally the mortality given that an individual has severe malaria) and 1— OR is the protection from malaria conferred by the genotype (after Hill, 1991 ; Hedrick, 2004 ). (Note that both Hill (1991) and Hedrick (2004) used the symbol RR in this equation, but in fact used the definition of OR given in equation (1a), reflecting the similarity of these measures for rare genotypes and the less specific use of these terms in earlier studies). For example, for the AS genotype if OR =0.082 as above and m =0.1 (see below), then s =0.092.
Hill (1991) suggested a value of 0.07 for m whereas from studies by both WHO (1998) and Carter and Mendis (2002) , a value of at least m =0.1 may be appropriate. However, recognizing that the appropriate value of m here should reflect the mortality in the evolutionary period over the many centuries when malaria was endemic and that it should account for the cumulative mortality of individuals with malaria before reproduction (and selection should include the impact on reproductive and mating success), an even higher value may be appropriate. For example, Rowe et al. (2006) found that in rural West Africans in whom malaria is hyperendemic and there is inadequate medical care, ∼ 20% of the children die of malaria by the age of 5. As a result, I will use m =0.1 here as a general value for illustration, with the caveat that m , and the consequent selection coefficient, may actually have been significantly higher in the past.
Because this selection gives an advantage to the AS genotype, the relative fitness of the AS genotype is the highest (1) and the relative fitness of genotype AA is 1— s or 0.908; therefore, the relative fitness levels of genotypes AA , AS and SS become 0.908, 1 and 0. From these values, the expected equilibrium frequency of allele S ( Hedrick, 2011a , p. 136) is 0.092/(0.092+1.0)=0.084, not unlike that observed in many sub-Saharan populations.
The age of two of the putative S alleles has been estimated by examining the extent of linkage disequilibrium surrounding the HBB locus. Currat et al. (2002) examined the S Senegal haplotype in a sample from eastern Senegal, in which there has not been recent admixture, and found that all haplotypes were identical. Using a selective advantage of 0.152 for AS genotypes over AA , they estimated that the age of S Senegal alleles was between 45 and 70 generations (1125 and 1750 years using 25 years per generation; see Table 2 ). Currat et al. (2002) suggested that the S mutation may have been at low frequency in the population and that this age estimate reflects the time since selection for malaria resistance increased this Senegal haplotype. Modiano et al. (2008) estimated the age of the Benin S haplotype by estimating the number of generations taken to decay to the observed linkage disequilibrium from the maximum expected when the S mutation occurred. They estimated that the age of the mutation was very recent and between 10 and 28 generations (250–700 years).
What is the expected future frequency of S alleles, because malaria has been eradicated in many areas and because modern medical care has increased the survival of individuals with sickle-cell anemia? First, heterozygotes AS do not appear to have symptoms of anemia or other effects (costs) of carrying the S allele in non-malarial environments ( Sheng et al., 2010 ), although some negative effects for AS individuals during strenuous exercise have been suggested ( Connes et al., 2008 ; Tsaras et al., 2009 ). As a result, the National Collegiate Athletic Association in the USA has instituted mandatory testing for sickle-cell carrier status for student athletes in its Division 1 sports ( Bonham et al., 2010 ). Second, with modern medical care, individuals with SS can live up to adulthood and reproduce with a much less reduction in fitness than without modern medical care. For example, Platt et al. (1994) found that the median age at death for individuals with sickle-cell anemia in the United States was 42 and 48 years for males and females, respectively (see similar results in Jamaica in Wierenga et al., 2001 ). As a result, in the absence of malaria and with modern medical care, the relative fitness levels of genotypes AA , AS and SS can generally be indicated by 1, 1 and 1— s . With these fitness levels, we can calculate how long it would take for selection (against SS homozygotes) to reduce frequency of the S allele from q 0 in generation 0 to q t in generation t using the following equation
( Hedrick, 2011a , p. 126).
In African-Americans, the frequency of S is about 0.05 and the proportion of individuals with sickle-cell anemia is about 1 in 400. How long would it take for the frequency of the S allele to be halved to 0.025 (this decreases the expected proportion of individuals with sickle-cell anemia to 1 in 1600)? With q 0 =0.05 and q t =0.025, the term in brackets in equation (4) is 20.7. Therefore, the number of generations to halve the frequency of allele S is about 20.7/ s . If we assume that s is still quite large, say 0.5, then it will take 41.4 generations, or 1035 years, to halve the frequency, quite a long time (of course if s is smaller, the decline would be even slower). The basis for this very slow rate of decrease is that most S alleles are in heterozygotes. Assuming that the frequency of alleles A and S are p and q , the proportion of S alleles in heterozygotes is pq/ ( pq + q 2 )= p. In other words, 95% of the S alleles in this example are in heterozygotes in which selection cannot act against them. Consistent with this prediction, Hanchard et al. (2007) found no significant difference between haplotype frequencies from Jamaica, where there has not been malaria since 1963 (about two generations), and the ancestral African haplotypes. This is in contrast to when selection favored S because of protection from malaria and selection acted favorably on the proportion p of the S alleles that were in heterozygotes and consequently resulted in a fast increase in allele frequency.
Gouagna et al. (2010) showed that there is higher transmission from the human host to the Anopheles mosquito vector of P. falciparum for S - and C -carrying individuals. This is an example of how host genetics can influence the efficiency of malaria transmission, but here the same host genotypes that confer protection against malaria appear to increase its transmission. However, individuals with these protective genotypes also use anti-malarial drugs less frequently because they may have more chronic, asymptomatic infections and therefore may become higher transmitters of infection ( Gouagna et al., 2010 ). Overall, it is not clear as to how these effects influence disease incidence and mortality in genotypically different individuals.
C or β C or HbC
Allison (1956) examined the effects and frequency of the hemoglobin C variant as well as sickle cell in his early studies in Africa. C is much more localized in central West Africa and reaches an allele frequency up to 50% in the Ivory Coast. C provides protection against P . falciparum malaria in both heterozygotes AC and homozygotes CC , although the estimated level of protection in heterozygotes appears to vary in different studies ( Modiano et al., 2008 see discussion below). The protective effect of C may result from increased immune clearance of infected erythrocytes. From examination of haplotypes and from its geographic distribution, there appears to be a single origin of C in Africa ( Flint et al., 1998 ). The C variant has also been found in Thailand on a different, non-African haplotype, indicating a second, independent origin ( Sanchaisuriya et al., 2001 ). Because in routine hemoglobin typing, C is often mistaken for variant E , which is common in parts of Thailand, the frequency of C in Thailand is not known precisely.
The change in the β-globin molecule for C is at the same amino-acid position as for S , but results in a change from glutamic acid to lysine rather than to valine (β6 Glu → Lys or GAG → AAG). There does not appear to be any cost from anemia or other traits to either the heterozygote AC or the homozygote CC , although CC homozygotes have symptoms comparable with a mild form of thalassemia. Individuals with the genotype SC have anemia, but it is milder than sickle-cell anemia (the much lower fitness of SC than AS indicates that C is different from the wild-type A allele). As a result, it appears that C could eventually go to fixation in malarial environments and could even replace S ( Modiano et al., 2001 ; Hedrick, 2004 ). However, Modiano et al. (2008) suggested that C is not as widespread as S because it is recessive in its advantage for malaria resistance (see below for a discussion).
Wood et al. (2005) used a coalescent-based approach to jointly estimate the time since the C allele began to increase in frequency because of strong selection and the selection coefficient utilizing data on the present allele frequency (0.15), effective population size and recombination rate. They estimated that the age of the allele was between 75 and 150 generations (1875 and 3750 years) and that the selection coefficient s was between 0.04 and 0.09 (assuming that the relative fitness levels of genotypes AA, AC and CC were 1, 1+ s , and 1+2 s , respectively). Modiano et al. (2008) calculated the age of the C haplotype in Burkina Faso by estimating the number of generations taken for linkage disequilibrium to decay to the observed level from the maximum expected when the C mutation occurred. They estimated that the age of the mutation was between 38 and 120 generations (950–3000 years), at the lower range of the estimate of Wood et al. (2005) .
E or β E or HbE
Hemoglobin E is the most common structural hemoglobin variant in Southeast Asia and has reached an allele frequency of up to 70% in some areas of northern Thailand and Cambodia. In general, neither hemoglobin S nor C variants are present in Southeast Asia and hemoglobin E is generally absent from populations in which S and C are present. This appears to be the result of historical differences resulting from mutational origin, but little is known about the interaction of E with S and E with C ( Fucharoen, 2001 ). It results from the substitution of lysine for glutamic acid in codon 26 (β26 Glu → Lys and GAG → AAG). Besides being a structural variant, the E variant also causes the production of an abnormal mRNA with less β-globin being synthesized. As a result, there is a mild thalassemia phenotype in EE homozygotes, but it does not appear to have an effect in AE heterozygotes (however, there is a much lower fitness of compound heterozygotes with both the E and the α- and β-thalassemia haplotypes; Fucharoen, 2001 ). AE heterozygotes appear to have protection from invasion into erythrocytes by P. falciparum malaria ( Chotivanich et al., 2002 ). Where E is in high frequency, some other red cell disorders, such as α-thalassemia, are also in high frequency.
Although extensive sequence analysis has not been carried out, from examination of the haplotypic backgrounds for E alleles in northeast Thailand, more than one mutational origin ( Fucharoen et al., 2002 ) is suggested. However, the E allele found in China is on the same haplotype as that found in Thailand ( Ohashi et al., 2005 ), suggesting that it does not have a different origin. From forward simulations, which used the amount of linkage disequilibrium close to the variant and the observed allele frequency, the age of the E allele was estimated to be about 100.3 generations ( Ohashi et al., 2004 ), or 2508 years, and the estimated selection coefficient between genotypes AA and AE was 0.079.
α – thalassemia
As I mentioned above, there are two linked genes with identical amino-acid sequences, HBA1 and HBA2 , on chromosome 16 that code for α-globin. There are two primary types of α-thalassemia: α + thalassemia, in which one pair of the genes is deleted or inactivated by a point mutation, and α 0 -thalassemia, in which both pairs of genes are deleted or inactivated. The deleted forms appear to be generated at a relatively high rate by unequal crossing over and Lam and Jeffreys (2006) estimated the rate of deletion generation in sperm from two men as 0.42 × 10 −4 . The haplotypes or gametes for the deleted forms with 2, 1 and 0 genes are indicated by αα, −α and −−, and the homozygous forms of α + thalassemia and α 0 thalassemia are indicated by −α/−α and −−/−−. The α + thalassemia is the most common monogenic disease in the world; however, as a homozygote, it results in an extremely mild anemia, −−/αα heterozygotes also have a mild anemia, similar to homozygous α + anemia, that is, the phenotypic effect appears to be the same whether the deletions are on the same chromosome −−/αα or on different chromosomes −α/−α. For heterozygotes with three α genes, or −α/αα, there is no anemia. When individuals are homozygous for deletions of both genes, −−/−−, or α 0 thalassemia, it is lethal and they are stillborn.
The frequency of α + thalassemia is generally >10% in regions where malaria is prevalent and in some populations, such as in Nepal, parts of India and Papua New Guinea, it is over 80% ( Flint et al., 1998 ). In fact, α + thalassemia appears similar to a transient polymorphism found in Nepal and Melanesia and it might eventually reach fixation given that selection continues to favor the −α haplotype. However, in sub-Saharan African populations, α + thalassemia frequencies do not exceed 50% despite intense malaria selection and Williams et al. (2005a) suggested that this might occur because of negative epistasis with the S allele (see discussion below).
The four different deletions that commonly cause α + thalassemia (out of about 80 in total) have different somewhat non-overlapping distributions. For example, deletion −α 3.7 is found nearly worldwide, but is most common in Africa, deletion −α 4.2 has a high frequency in Southeast Asia and −α 3.7I is found most commonly in African, Indian and Mediterranean populations ( Flint et al., 1998 ). In addition, there are many different local forms of these single-gene deletions, as well as of the deletions that eliminate both genes. Although the age of these variants has not been estimated, their localized presence suggests that they are recent, have arisen in different malarial areas and are increasing because of their selective advantage.
The distribution of both α and β thalassemia variants correspond closely to the regions that have historically had high rates of malaria ( Weatherall and Clegg, 2001 ) and the local distribution of these variants also corresponds to endemic malaria ( Siniscalco et al., 1961 ; Flint et al., 1986 ; Hill et al., 1988 ). In addition, several studies have shown protection from severe malaria for individuals with α + thalassemia, compared with individuals without thalassemia ( Allen et al., 1997 ; Mockenhaupt et al., 2004 ; Williams et al., 2005b ). In a large case–control study in Kenya ( Williams et al., 2005b ), the OR values, standardized to the normal αα/αα genotype, for α + heterozygotes and homozygotes in Kenya are 0.623 and 0.522 for severe malaria, respectively ( Hedrick, 2011b ). In the other case–control study in Papua New Guinea ( Allen et al. 1997 ), the standardized OR values of α + thalassemia heterozygotes and homozygotes are 0.781 and 0.362, respectively, for severe malaria. Overall, it appears that many haplotypes that reduce expression of α-globin provide a selective advantage in resistance to severe malaria. Such a general advantage to an entire class of variants that reduces expression appears to be also true for β-globin variants and for G6PD-deficiency variants that reduce enzyme expression (see below).
Using the OR values mentioned above and equation (3), and assuming that m =0.1, the estimated relative fitness levels of individuals with different genotypes with different numbers of functional α gene copies are given in Table 3 . Here, the fitness of −−/−− is 0, as discussed above and the fitness of −−/−α with one gene is w 1 and the fitness of −−/αα and −α/−α individuals are the same.
From these values, I intuitively expected the −α gamete to increase in frequency to fixation, except for unequal recombination generating other gametes, and for there to be strong selection against −− gametes. This is true for the estimated Kenyan relative fitness values mentioned above for all values of w 1 ( Hedrick, 2011b ). However, for the Papua New Guinea fitness values −α goes to fixation only when genotype −−/−α has a fitness of 0.79 or above. When w 1 is 0.78 or less, haplotype −α does not increase in frequency and haplotypes −− and αα go to a heterozygote advantage polymorphic equilibrium, not substantially different from that observed for some Asian populations ( Hedrick, 2011b ).
β–thalassemia
Remember that the ‘malaria hypothesis’ of Haldane (1949b) was proposed to explain the very high level of β-thalassemia in some Mediterranean populations. There is ordinarily only one copy of the HBB gene and β + and β 0 thalassemia indicate the reduction and loss, respectively, of the production of functional protein. Individuals with β-thalassemia major are often homozygous for loss of production of β-chain mutants (although they may be compound heterozygotes for two mutants), have profound anemia and, if they are not treated with blood transfusions, die in their first year. Heterozygotes typically have mild anemia (β-thalassemia minor), but symptoms can vary greatly in severity from having severe anemia to being a symptomless carrier. In general, β-thalassemia is more of a public health problem because of this higher morbidity than α-thalassemia.
Most of the β-thalassemias are the result of single-nucleotide substitutions or small insertions or deletions. Around 200 are known worldwide with many different local forms; therefore, the fact that they are the result of different mutations appears clear. Also, because of the local distributions, it appears that the mutants are recent in age. The frequency of carriers of β-thalassemia variants is from 5 to 20% in some areas, although not as high as the frequency of α-thalassemia variants ( Weatherall and Clegg, 2001 ).
Worldwide distribution of β-thalassemia generally coincides with historic distribution of malaria. Malaria is not present today in most areas with higher frequencies of β-thalassemia, such as around the Mediterranean; therefore, case–control studies are not possible. However, Willcox et al. (1983) carried out a case–control study in an endemic malaria area of Liberia and found evidence for malaria protection in β-thalassemia heterozygotes.
β-thalassemia variants exist in areas where there are structural β-globin variants and as a result, compound heterozygotes (heterozygotes with two different mutants) occur. For example, in areas of Africa and around the Mediterranean, there are S β-thalassemia heterozygotes. The severity of disease in these individuals varies depending on the nature of β-thalassemia allele, but at its most extreme can resemble sickle-cell anemia. In Southeast Asia, where the structural variant E is common, the compound heterozygote E β-thalassemia is also common ( O'Donnell et al., 2009 ) and causes about 1/3 of the cases of severe thalassemia in Sri Lanka.
Enzymopathies
G6pd deficiency.
Glucose-6-phosphate dehydrogenase, G6PD, is an important enzyme that catalyzes the first reaction in the pentose-phosphate pathway, part of glycolysis ( Cappellini and Fiorelli, 2008 ). In the erythrocyte, G6PD is the sole source of enzyme activity that protects from the buildup of super-radicals and to withstand oxidative stress. G6PD deficiency is the most common enzymopathy in humans, affecting around 400 million people worldwide with a global prevalence of 4.9%, although there is substantial variation among populations ( Nkhoma et al., 2009 ). Although G6PD-deficient individuals are generally asymptomatic throughout their life, factors that cause oxidative stress, such as particular drugs (including several antimalarials), foods or infections, can cause hemolytic anemia and neonatal jaundice. For example, digestion of fava beans can cause severe hemolytic anemia in individuals with the Med G6PD deficiency, a condition traditionally known as favism. Low mortality and low morbidity are associated with G6PD deficiency and it does not seem to generally affect life expectancy.
The G6PD gene has 13 exons, is 18 kb in length, is on the X chromosome close to the genes for hemophilia and color blindness and codes for a molecule that is 515 amino acids long. The active enzyme is composed of two or four identical 515 amino-acid subunits. The G6PD gene is one of the most polymorphic loci in the human genome and about 140 different molecular variants have been identified ( Cappellini and Fiorelli, 2008 ). Most of these have non-synonymous single-nucleotide replacements, a number of which are polymorphic. These variants range from ones that cause severe deficiency and chronic anemia to some that actually increase G6PD activity. G6PD deficiency can be caused by a reduction in the number of enzyme molecules or a structural change causing enzyme instability. Because G6PD is on the X chromosome, males with a single deficient G6PD allele are affected. For females to show deficiency, they generally need to be homozygous for the deficient variant, although some heterozygous females can exhibit G6PD deficiency and even develop hemolytic anemia.
The ancestral normal activity G6PD allele B is present worldwide, but the variants causing G6PD deficiency are generally more localized to specific geographic areas. Similar to the sickle-cell allele, the worldwide historic distribution of G6PD-deficient mutations reflects the historic distribution of malaria endemicity. For example, much of Africa has three main polymorphic alleles, the ancestral B allele with a frequency of about 0.6–0.8, the A allele, which differs from B by one amino acid (resulting from a difference at one nucleotide) and has a frequency of about 0.15–0.4, and the A - allele, which differs from A by one amino acid (also resulting from a difference at one nucleotide) and has a frequency ranging from 0.0 to 0.25 ( Table 4 ).
The A allele has 85% of normal G6PD activity and has not been demonstrated to provide resistance to malaria in contemporary populations. On the other hand, the A - allele has only 12% of normal activity and is thought to provide resistance to malaria (the A - allele is always found on the A background, so this combination may be necessary for malaria protection). The Mediterranean or Med allele is the second most common allele found in countries surrounding the Mediterranean Sea, India and Indonesia. It differs from allele B by one amino acid (resulting from a one-nucleotide change) at another site from that of A , has only 3% normal activity and is in frequencies ranging from 0.02 to 0.2. The Mahidol allele also differs from B by one amino acid (also from a one-nucleotide change) at another site, has 5–32% the activity of the normal enzyme and reaches a frequency of 0.24 in some populations in Southeast Asia.
Overall, it appears in general that a reduction in G6PD enzyme activity, due to these variants and others, results in an increase in resistance to severe malaria. This is not unlike the general effect from number of variants at the α-globin and β-globin genes that result both in thalassemia from lowered gene expression and consequently an increase in resistance to severe malaria. The exact mechanism of increased resistance to malaria from G6PD deficiency is not known, but appears to be related to the inability of the malaria parasite to survive and reproduce in stressed G6PD-deficient cells.
To understand the effect of X-linkage on G6PD frequencies, let me assume that the frequency of a G6PD deficiency allele, say A - is q (and the frequency of the non-deficiency allele is 1− q ). Because males are hemizygous for genes on the X-chromosome, the frequency of males with G6PD deficiency is equal to the frequency of the A - allele, or q . Assuming Hardy–Weinberg proportions, the three female genotypes, BB , BA - and A - A -, have frequencies of (1− q ) 2 , 2 q (1− q ) and q 2 , respectively. All female A-A - individuals have G6PD deficiency, but at a rate that is only q as much as males. However, some percentage of females heterozygous for G6PD-deficient alleles also show deficient G6PD activity, making this reduction somewhat less. On the other hand, if DNA analysis is used to detect the deficiency allele in heterozygotes, regardless of whether they show G6PD deficiency, then the proportion of females with the deficiency allele, either as heterozygotes or homozygotes, is 2 q (1− q )+ q 2 =2 q − q 2 , a value that is nearly twice as high as the frequency in males.
The results of studies examining the risk of malaria for various G6PD-deficient genotypes are not consistent. For example, Ruwende et al. (1995) found in Gambia and Kenya that the reduction in risk (1— OR ) of severe malaria in male hemizygotes ( A -) was 58% and that the reduction in risk for heterozygous females ( BA -) was 46%. On the other hand, Guindo et al. (2007) found that in two populations in Mali, the reduction in risk of severe malaria in male hemizygotes ( A -) was also 58%, but in female heterozygotes ( BA -) there was no reduction in risk. Further, when Guindo et al. (2007) examined only those individuals who were wild-type AA at the β-globin locus, the effect in males was increased ( OR =0.28) whereas there was still no effect in females. Guindo et al. (2007) suggested that the difference in their results and that of Ruwende et al. (1995) might be based in the control groups used. In addition, Johnson et al. (2009) found no protective effect for either male hemizygotes ( A -) or female heterozygotes ( BA -) in a Uganda population, but did find a protective effect for females that were found to be G6PD deficient. This perplexing finding appears to be based on the incomplete correlation of genotype and phenotype for G6PD deficiency in female heterozygotes due to variable inactivation of the two X chromosomes ( Mason et al., 2007 ).
If we use the OR values from Ruwende et al. (1995) and the approach mentioned above to estimate selection coefficients in females ( s f ) and in males ( s m ), then they are 0.046 and 0.058, respectively. The three female genotypes, BB, BA and A-A- have fitnesses of 1– s f , 1 and 1, respectively, assuming that the heterozygote and the mutant homozygote have the same fitnesses (see Saunders et al., 2002 ). The two male genotypes, B and A -, have fitnesses 1 –s m and 1, respectively. Therefore, if we assume that the initial frequency of A - is 0.01, and using traditional equations for change in allele frequency for X-linked genes ( Hedrick, 2011a , p. 154), A - increases quickly and reaches a frequency near 1 by 200 generations (solid line in Figure 1 ). Interestingly, the examples considered in all three figures here (G6PD deficiency, HBC and α + thalassemia) include situations in which a malaria-resistant variant could go to fixation. If there is a symmetrical heterozygote advantage in females because the A - A - homozygotes have a lowered fitness due to G6PD deficiency, and no differential selection in males, then the frequency approaches its equilibrium value of 0.5 in about 300 generations (broken line in Figure 1 . When the A - A - females and the A - males have a selective disadvantage, but there still is a heterozygote advantage in females, then the frequency of A - slowly approaches its equilibrium value of 0.126 (dotted line in Figure 1 ), a value not too different from that observed in many populations.
The increase in frequency of allele A - at the G6PD locus when it begins at a frequency of 0.01 and fitness of females BB , BA - and A - A - and males B and A - are for females 1— s f , 1, and 1 and for males 1— s m and 1 (solid line), for females 1— s f , 1, and 1— s f and for males 1 and 1 (broken line), and for females 1— s f , 1, and 1—2 s f and for males 1 and 1 - s m (dotted line), where s f =0.046 and s m =0.058 (estimated from the data of Ruwende et al., 1995 ).
The age of G6PD mutants have been estimated using several different approaches, some of which assume selection (assuming s f = s m = s ) and others do not ( Table 2 ). Tishkoff et al. (2001) jointly estimated the age and selection for both A - and Med alleles using forward simulations and found that the estimated age of A - was 6357 years (254 generations) and the selection coefficient was 0.044, and that the estimated age of Med was 3330 years (132 generations) and the analogous selection coefficient was 0.034. From joint estimations using Bayesian methods, Slatkin (2008) estimated that the age of A - was only 1000 years (40 generations) and the selection coefficient was 0.25 and Louicharoen et al. (2009) estimated that the age of allele Mahidol was 1575 years (63 generations) and the selection coefficient was 0.23. The difference in these estimates may be because they were based on different data, with that of Tishkoff et al. (2001) based on microsatellite data and that of Slatkin (2008) based on sequence data. The selection coefficients and consequent times in Figure 1 are somewhat more consistent with that estimated by Tishkoff et al. (2001) than the estimates of Slatkin (2008) and Louicharoen et al. (2009) .
Carter and Mendis (2002) suggested that the original selective force for G6PD deficiency could have been either for resistance to P. falciparum or P. vivax . As evidence for the importance of P. vivax , they cite the high frequency of G6PD deficiency in a population in northern Holland, an area where P. vivax was prevalent for centuries and P. falciparum is thought to have never existed. Recently, evidence has suggested that G6PD deficiency caused by the Mahidol mutation in Southeast Asians provides parasite resistance to infection by P. vivax , but not to that by P. falciparum . Further, G6PD deficiency Med in Pakistan has been shown to provide protection from P. vivax malaria ( Leslie et al., 2010 ).
Red blood cell surface loci
Two red blood cell loci, Duffy and ABO, that appear to provide protection from malaria, are similar in one sense that they both have two common functional alleles and a defective, loss-of-function allele that consequently provides protection against malaria. On the other hand, the Duffy variant protects against P. vivax , whereas the O phenotype of the ABO system appears to provide protection against P. falciparum . The gene for band 3 ovalocytosis, the major transmembrane protein of red blood cells, also has a deletion, loss-of-function, mutant. This mutation provides protection from cerebral malaria, is lethal in homozygotes and appears to provide protection from both P. vivax and P. falciparum .
The Duffy blood group antigen was first observed in 1950 and has two common antigens A and B produced by alleles FY*A and FY*B (for a review of the Duffy gene and malaria, see Zimmerman, 2004 ). The Duffy gene is symbolized either by FY or DARC (Duffy antigen receptor for chemokines). The FY*B allele is in highest frequency in Europe whereas the FY*A allele is nearly fixed in some Asian populations. Surveys showed that many African-Americans and Africans did not possess either the A or B antigens and had another allele, a Duffy-negative allele that did not produce antigens on erythrocytes and is known generally as a Duffy ‘null’ allele ( FY*A and FY*B are also called Duffy positive). This allele, FY * B ES , now termed erythrocyte silent (ES), is near fixation in sub-Saharan Africa (it is not a true null allele because it is expressed on cell types besides erythrocytes). From analysis of the sequences of these alleles, it appears that FY*A is ancestral with amino acid Gly (TGG) at codon 42. Allele FY*B differs from FY*A with Asp (TAG) at position 42. The ES allele in Africa differs from FY*B with a T to C SNP mutation, 33 nucleotides upstream from the transcription starting point (−33). This mutation disrupts a binding site for transcription factor GATA-1 on the gene promoter and blocks expression of the gene in erythrocytes.
Several experiments showed that Duffy ES homozygotes ( FY * B ES /FY * B ES ) were resistant to infection by two malarial species, P. knowlesi and P. vivax , but were susceptible to infection by P. falciparum ( Miller et al., 1976 ). From these and other studies, it was shown that the Duffy antigen was the obligatory binding receptor used by P. knowlesi and P. vivax to enter red blood cells and that in homozygous ES individuals, no antigen was present on red blood cells and these malaria species could not infect them. In fact, it has been proposed ( Hamblin and Di Rienzo, 2000 ) that the near absence of P. vivax in Africa, which is thought to have originated in Asia, occurs because there were no suitable hosts in Africa even though the African environment appears otherwise suitable for P. vivax . That is, nearly all Africans were FY * B ES /FY * B ES , because of an earlier selective sweep due to some previous selective force when P. vivax moved west to Africa from Asia. On the other hand, selection for resistance to P. vivax in Africans could have increased the frequency of the FY * B ES allele and subsequently eliminated P. vivax from Africa ( Carter and Mendis, 2002 ).
There does not appear to be any cost (or disease) associated with the presence of FY * B ES homozygotes or heterozygotes. If there were a cost, then because there is no P. vivax in Africa and therefore no selection for the FY * B ES allele, it would be expected that the frequency of FY * B ES would decline. Similarly, the FY * B ES allele frequency in African-Americans would not be expected to decline, even in the absence of P. vivax malaria, unless there was a pleiotropic cost to maintaining the allele.
Analysis of the population genetic aspects of the Duffy alleles in Africa is consistent with strong past selection. For example, the F ST value for the FY * B ES allele is one of the highest observed for any alleles in humans ( Hamblin and Di Rienzo, 2000 ; Hamblin et al., 2002 ). Further, the level of variation around the FY * B ES allele in Africa is significantly lower than that observed in an Italian sample for the Duffy region. Interestingly, the FY * B ES allele in Africa is found on two different haplotypes in three of the five African populations examined by Hamblin and DiRienzo (2000), suggesting either two mutational origins of the FY * B ES in Africa or recombination/gene conversion between these haplotypes.
On the basis of phylogenetic relationships of the FY sequences, Hamblin and Di Rienzo (2000) estimated the age of the FY * B ES allele around 33 000 years ago. As P. vivax may have been a significant cause of morbidity before human populations increased in density because of the development of agriculture around 5000 to 10 000 years ago, they suggested that this earlier origin may be consistent with the suggested earlier impact of P. vivax . Using a microsatellite locus linked to FY , Seixas et al. (2002) estimated that the origin of the FY * B ES allele was more recent, either 7750 or 12 250 years ago, depending upon the haplotype they used (this assumes a generation length of 25 years).
Surprisingly, a Duffy ES allele found in low frequency (0.022) in Papua New Guinea, differed from FY*A in exactly the same way at the −33 nucleotide in the promoter region as does the FY * B ES allele found in Africa ( Zimmerman et al., 1999 ) and is now called FY * A ES . Although this allele appeared to be on a FY*A background from analysis of restriction fragment patterns, more detailed sequence analysis could definitively exclude African ancestry for this allele. Zimmerman et al. (1999) also documented that there is half as much expression in these heterozygotes FY*A / FY * A ES as that in normal homozygotes FY*A / FY * A . Further, these heterozygotes were protected from P. vivax infection more than normal homozygotes whereas they were not protected from P. falciparum infection ( Kasehagen et al., 2007 ). The low frequency of FY * A ES appears to be the result of its recent origin, which is reflected in its association with specific alleles at two microsatellite loci about 3 cM from FY ( Zimmerman et al., 1999 ). Recently, Ménard et al. (2010) also showed that there is about half as much expression in a sample from Madagascar for FY*A / FY * B ES heterozygotes as that for for normal homozygotes ( FY*A / FY * A ) or heterozygotes ( FY*A / FY * B ). In other words, it does not appear that either the expression or the protection from ES alleles is recessive, but is intermediate between ES homozygotes and genotypes with two normal alleles.
Recently, there have been reports of P. vivax in FY * B ES individuals from Brazil, Kenya and Madagascar. Perhaps the most definitive study is that of Ménard et al. (2010) in Madagascar, a nation where endemic P. vivax is prevalent and a diverse ethnic population with both Duffy-negative ( FY * B ES /FY * B ES ) and Duffy-positive (having at least one allele that is FY * A or FY * B ) individuals. In the sample examined of 476 asymptomatic Duffy-negative individuals from eight locations, P. vivax was found in 8.8%. However, they still found a substantial reduction in clinical P. vivax malaria among Duffy-negative individuals compared with Duffy-positive individuals. It appears that this population with both Duffy-negative and -positive individuals provided an environment for strains of P. vivax to evolve the ability to invade Duffy-negative individuals, giving an example of evolution for utilization of an alternate receptor besides the Duffy antigen for erythrocyte invasion.
SLC4A1 or band 3 ovalocytosis
Gene SLC4A1 (also known as erythrocyte Band 3 protein gene) codes for the major transmembrane protein of red blood cells and is an anion exchanger in the erythrocyte membrane. One mutation in this gene, found in Papua New Guinea and Malaysia, is due to a 27-nucleotide (9-amino-acid) deletion in a region that is highly conserved throughout species ( Jarolim et al., 1991 ). This deletion results in an abnormal protein structure and function and causes ovalocytosis, an abnormal shape of red blood cells (it is known as Southeast Asian ovalocytosis, or SAO), and mild hemolytic anemia in the heterozygote. The homozygote appears to be lethal because it has never been observed even in progeny from heterozygous matings ( Genton et al., 1995 ).
In populations from lowland Melanesia with high malaria rates, the frequency of this deletion can be as high as 15–20%, but it is not present in the malaria-free highlands. Neither S nor E hemoglobin variant is present in these populations and only some thalassemias and G6PD deficiency are known to be present to reduce the effects of malaria. Because most individuals in many of these areas are infected with malaria, and often multiple malaria species ( Mueller et al., 2009 ), other unidentified resistance variants appear to be present. There is substantial evidence that deletion heterozygotes are highly protected against malaria. For example, the frequency of the deletion was 0.146 in a sample of healthy Papua New Guinea children whereas it was not present in any of the children with cerebral malaria ( Genton et al., 1995 see also; Allen et al., 1999 ). Carriers also appear to be at lower risk of infection by both P. vivax and P. falciparum ( Cattani et al., 1987 ; however, see Kimura et al., 2002 ).
The lethality of homozygotes and the high resistance of heterozygotes to malaria suggest that this deletion variant is somewhat similar to the sickle-cell allele in that the benefit to heterozygotes is high enough to counter the large fitness cost in homozygotes. However, it appears to be a relatively recent mutation because of its limited distribution and a detailed examination of variation at closely linked sites may provide insight as to its age.
For many decades, the ABO blood group system has been suggested to be associated with diseases and infections, including malaria. The gene consists of seven exons, is more than 18 kb in length and genomic analysis has found over 70 alleles at this locus, suggesting that it is one of the polymorphic genes in humans ( Calafell et al., 2008 ). The ABO glycosyltransferase performs the final step in the production of the ABO molecule by adding sugars to the precursor H antigen. This enzyme either adds the sugars N-acetylgalactosamine or galactose to form the A and B antigens or does not add a sugar, resulting in a functionless H (O) antigen. The three main antigenic classes (A, B and O) are all comprised of numerous alleles determined by both coding and non-coding sequences ( Calafell et al., 2008 ). The silent O alleles share a one-nucleotide deletion in codon 87 (nucleotide 261) of exon 6 that results in a frameshift mutation and premature termination of the polypeptide ( Yamamoto et al., 1990 ). The O alleles are the most common of the three allelic classes (about 0.6 worldwide) and have frequencies between 0.3 and 0.7 in most populations, the A alleles generally have frequencies between 0.2 and 0.3 and the B alleles have frequencies between 0.1 and 0.2. Native American or Australian aboriginal populations have O allele frequencies of nearly 1.0 ( A and B do not appear to have not been present historically in these populations).
Cserti and Dzik (2007 ) suggested that there is substantial evidence supporting the importance of allele O providing protection against malaria. Their conclusion is based primarily on the consistency between the worldwide distribution of ABO variants and historic presence of malaria, with the O allele more common in areas with historic malaria and the association of O genotypes with higher resistance to malaria and A and B genotypes with lower resistance in a number of studies ( Uneke, 2007 ). In a large recent study, Fry et al. (2008) showed in three African populations a strong association of O individuals with resistance to severe malaria and found that this effect was recessive, that is, AO and BO individuals were as likely to be susceptible as AA and BB individuals and that AB genotypes were the most susceptible. In addition, it appears that the O allele can protect against severe malaria by a mechanism of reduced rosetting (spontaneous binding of infected erythrocytes to uninfected erythrocytes) ( Rowe et al., 2007 ).
Fry et al. (2008) calculated F ST values for the ABO genomic region and found that, even with its polymorphism within populations, it was an outlier for low F ST over populations. They suggested that this similar variation over different populations is the result of similar long-standing balancing selection over populations, perhaps caused by infectious pathogens, including P. falciparum . However, Calafell et al. (2008) found a more complicated pattern of differentiation over the ABO genomic region. Although the major O alleles share a one-nucleotide deletion, they differ in a number of nucleotide substitutions in both exons and introns. Unlike the other recent malaria-resistant alleles discussed here, the O human alleles, although different from the O chimpanzee allele ( Kermarrec et al., 1999 ), are much older. Calafell et al. (2008) estimates that the most common O alleles, O01 and O02 , are the result of separate mutations and are ∼ 1.15 and 2.5 million years old, respectively. In other words, assuming that the O allele provides protection from malaria, this protection may be present because of selection originally favoring O alleles for some other reason.
Immune genes
The genes I have discussed above, such as the hemoglobin mutants and G6PD deficiency, provide innate resistance to malaria. In addition, although human immunity to malaria is complex ( Langhorne et al., 2008 ), variants in the HLA complex have been shown to provide adaptive immunity to malaria. In general, innate resistance is more important in early childhood survival from malaria whereas adaptive immune response is more important in older children and adults. It appears that malaria protection from particular HLA variants is local and not universal ( Hill et al., 1994 ; Ghosh, 2008 ). Various factors, such as variation over different populations in malaria strains (and species), in red blood cell polymorphisms, and in HLA polymorphism, may contribute to this heterogeneous response to malaria ( Ghosh, 2008 ).
HLA-Bw53 and DQw5
HLA genes are in the human MHC (major histocompatibility complex) and are the most variable genes in the human genome. They have been shown to be, and have been, under balancing selection, using a number of different approaches ( Garrigan and Hedrick, 2003 ). It is widely accepted that the mechanism of balancing selection at HLA, and for MHC genes in other vertebrates, which retains genetic variants at these genes over species (trans-species polymorphism), is related to the protection that they provide against infectious disease ( Hedrick and Kim, 2000 ). HLA (and MHC) genes can be divided into class I and class II genes in which class I genes recognize intracellular antigens, as from viruses or intracellular stages of parasites, and class II genes recognize extracellular antigens, as from bacteria and parasites. One of the first conclusive demonstrations that HLA variation provided resistance to contemporary infectious disease was the large study showing protection from both specific classes I and II HLA types to malaria in Gambia ( Hill et al., 1991 ).
As there are so many variant forms at many HLA genes (class I genes HLA-A and HLA-B have over 250 and 500 alleles, respectively; Garrigan and Hedrick, 2003 ), particular associations may have occurred by chance and after correction for multiple comparisons, statistical significance for particular associations may be lost. As a result, Hill et al. (1991) used a two-stage strategy in which they first examined all 45 possible class I HLA-malaria associations in approximately half their samples serologically, and then in stage two in an independent sample examined explicitly the strongest association found in the first group (Bw53), using PCR. As shown in Table 5 , the proportions of young children heterozygous for Bw53 were nearly identical in the two independent samples for both the severe malaria and the mild control categories (the mild control group consists of children attending the same hospitals or clinics as those with severe malaria matched for age and area of residence). The calculated OR values were nearly identical in the two samples and indicated that Bw53 confers substantial protection from severe malaria in this population. In addition, a class II haplotype (DQw5), consisting of both DQ and DR genes, also showed a strong protective effect in this population ( Table 5 ).
The frequency of individuals with Bw53 (either homozygotes or heterozygotes) is highest in sub-Saharan Africans, reaching 40% in Nigeria and 25% in Gambia, whereas it is very rare (<0–1%) in Asians and Caucasians. Therefore, not only does there appear to be a strong contemporary protective effect, but also its geographical distribution is not unlike that for the sickle-cell allele and indicates past selection favoring Bw53 in malarial environments. Similarly, haplotype DQw5 showed a novel arrangement of genes and it appears that this haplotype is not found, or is very rare, in Caucasians. Because of the high variation in the HLA region, it is not possible to age Bw53 in the way discussed above, but we can estimate the amount of selection and determine the number of generations it would take the genotype to increase in frequency to the present level.
For example, for the Bw53 genotype if OR =0.59 (the average of the values in Table 5 ) and m =0.1 ( WHO, 1998 ), then s =0.041 from equation (3). Using this amount of selection (and standardizing such that the relative fitness of genotype with the highest fitness is 1) and assuming that the Bw53 allele is dominant, the relative fitnesses of genotypes Bw53/Bw53, Bw53/B(not w53) and B(not w53)/B(not w53) are 1, 1 and 0.959, respectively, where B(not w53) indicates all other alleles at gene HLA-B . If we assume that the Bw53 was at an initial frequency of 0.005, as it is now in parts of Europe ( Hill, 1991 ), then using a traditional population genetics selection equation ( Hedrick, 2011a , p. 118), it would take ∼ 86 generations or 2150 years for the frequency of individuals with Bw53 to reach 0.25 ( Hill, 1991 ). This time is substantially less than the time that was expected for strong selection to have occurred for malaria resistance in Africa.
It appears that Bw53 was generated by a gene conversion event in which only seven adjacent amino acids of the most common allele in Gambia, Bw35, were converted to another common motif ( Allsopp et al., 1991 ). Because of the presence of potential donor sequences and the recipient sequence (Bw35) in Gambia, it appears that Bw53 arose locally. As the Bw35 allele does not appear to confer protection from malaria, it appears that this small converted region in its new background provides protection. Further examination of DQw5 by Davenport et al. (1995) showed that the higher resistance of this haplotype appeared to be the result of a one-amino-acid difference at the DRB1 gene and proposed that the resistance from the DQw5 haplotype occurs because this haplotype recognizes an epitope from the malaria parasite.
Multiple resistance variants in the same population
As we find out more information about genes conferring resistance to malaria, we can apply population genetics approaches to predict the eventual outcome of selection when there are two or more variants undergoing selection in a given population. Potentially, different resistance variants may be different alleles at a given locus in which selection may function along with the normal ancestral allele to increase, decrease or stabilize genetic change. Different resistance variants at different loci may be statistically associated (in linkage disequilibrium), resulting in change that is not independent over loci. Moreover, resistance variants at different loci may have fitness values that are not independent over loci (epistasis) and influence genetic change.
There are a number of known genes that could contribute independently (without epistasis) to malaria resistance in areas with high rate of infection; for example, G6PD deficiency and S in sub-Saharan Africa, and Duffy, α-thalassemia and ovalocytosis in Papua New Guinea. All of these genes are unlinked and are present on different chromosomes; therefore, no linkage disequilibrium between the resistance variants at different loci is expected, although there may be strong epistasis between them. However, as more resistance genes become known, some may be linked, so that both linkage disequilibrium and epistasis may become important factors influencing genetic change and overall resistance.
Here, I give two examples, one in which the genetic variants are different alleles at the same gene ( S and C ) and the other in which the genetic variants are different alleles at different genes, but there is epistasis between them ( S and α-thalassemia). Remember that the α-globin and β-globin molecules each contribute two subunits to adult hemoglobin molecules, suggesting that this essential connection could provide potential for gene–gene interaction, or epistasis, as far as fitness. The geographical distributions of α and β thalassemia overlap and in individuals with both, the more severe forms of β + thalassemia may be changed to a milder condition. Unbalanced production of α and β chains is a major cause of thalassemia, but if both are low, then the severity of anemia can be less. As a result, α-thalassemia can modify the phenotype of β-thalassemia such that it is not as extreme, a potential example of positive epistasis ( Penman et al., 2009 ). In addition, the sickle-cell anemia can be ameliorated by an increase in the production of fetal hemoglobin ( Flint et al., 1998 ), suggesting positive interaction between these genes.
Before I discuss the specific examples, let me provide a general perspective of resistance when several genes are involved. When several different resistance genes are present in the same population, assuming that the resistance variants function independently, the overall expected mean proportion of the population that is resistant to malaria R can be expressed as
where f ij and R ij are the frequency and resistance, respectively, of genotype j at the i th locus (notice that different loci may have different levels of dominance in this general equation). For example, let me assume that there are two resistant genotypes at locus 1 with frequencies f 11 =0.1 and f 12 =0.5 and resistances R 11 =0.5 and R 12 =0.3, and one resistant genotype at locus 2 with f 21 =0.3 and R 21 =0.5. Therefore, the mean proportion of resistance in the population is
or 32% of the population would be expected to be resistant to malaria.
The polymorphic structural variants at the β-globin locus, S , C and E , confer resistance to malaria. Variant C is more localized in sub-Saharan Africa than S , but does co-occur with S in many populations. For example, Livingstone (1967) complied survey data on 72 populations from West Africa (about 33 000 individuals) and found a negative correlation between the frequencies of S and C when both genes were present. In general, there were more populations with frequencies of C between 0.075 and 0.15 and S <0.05 and with frequencies of S between 0.075 and 0.15 and C <0.05 than expected. Although some early estimates of fitness suggested that there might be a stable equilibrium for the three alleles A , S and C , a very large data set from Burkina Faso from Modiano et al. (2001) suggested that the C allele would eventually become fixed in regions of malaria (see also Hedrick, 2004 ; Modiano et al., 2008 ). Below I will discuss this example and estimate relative fitness levels of the six genotypes when there is simultaneous segregation for S and C in the same population and then use these values to predict change in allele frequencies in this population.
Assume that genotype i provides protection from malaria because the frequency of genotype i in the diseased group is less than that in the control group, or that f id < f ic , as I did above. Further, if s i is the selection coefficient for genotype i , it can be estimated as in equation (3). Because selection for multiple genotypes at the locus is considered here (and relative fitness needs to scaled symmetrically around 1), w i , the relative fitness of genotype i , is
For example, if f id =0.2 and f ic =0.4 and m =0.5, then s i =0.3125 and w i =1.4545.
Now assume that f id > f ic , that is, genotype i provides relative susceptibility to malaria. In this case, the selection coefficient can be defined as
Because selection gives a disadvantage to the genotype, the relative fitness of genotype i is
For example, if f id =0.8 and f ic =0.6 so that OR =2.667. Further, if m =0.5, then s i =0.3125 and w i = 0.6875. Note that 1/0.6875=1.4545, or that the fitness levels in these examples are symmetrical around 1.
In the study by Modiano et al. (2001) , there were statistically significant effects for the hemoglobin genotypes AA , AC , AS and CC ( AC , AS and CC showed relative resistance and AA showed relative susceptibility). Table 6 lists the frequencies of these four genotypes in the control (healthy subjects) and disease (malaria patients) groups and the resulting OR values. The sample sizes for genotypes SC and SS were very small and the fitness levels used here were based on deviations from Hardy–Weinberg proportions ( Hedrick, 2004 ). Assuming m =0.1, Table 6 lists the relative fitness for these genotypes and also the fitness levels relative to the genotype with highest fitness, CC . Using these fitness levels, there are no stable or unstable three-allele equilibria, unlike earlier data ( Hedrick, 2004 ).
With this fitness level, if S is introduced by mutation or gene flow into a monomorphic A population, it will quickly go to a stable two-allele equilibrium with the equilibrium frequency of S equal to 0.12. If C is introduced by mutation or gene flow into a monomorphic A population, it will increase in frequency to fixation in slightly over 100 generations ( Figure 2 )). When the S and A alleles are at their stable equilibrium, C can enter the population and eventually increase to fixation after about 200 generations. When both the C allele and the S allele are introduced simultaneously at low frequencies (0.01 here), the S allele will be eliminated (in about 100 generations in the example in Figure 2 ).
The increase in frequency of allele C when it begins at a frequency of 0.01 (short, broken line), when S also begins at a frequency of 0.01 (solid lines), and when S begins at its equilibrium frequency of 0.0895 (long, broken line) (after Hedrick, 2004 ). The change in frequency of S is also given for the last two situations.
These outcomes occur primarily because genotype CC has the highest estimated relative fitness of any genotype. It may also explain particular situations as, for example, why the Dogon people of Mali have a much lower frequency of S than most other West Africans and have a high frequency of C ( Agarwal et al., 2000 ). Of public health significance, the mean fitness of the population will be 11.2% higher when the population is fixed for C than when it is polymorphic for A and S because of higher average resistance to malaria and the absence of sickle-cell anemia.
Modiano et al. (2008) suggested that C is not as widespread as S because it is recessive in its advantage for malaria resistance. To estimate dominance for the favorable C allele, we can standardize the fitness levels so that genotypes AA , AC and CC have fitness levels of 1, 1+ hs and 1+ s , respectively, where h is the level of dominance for the favorable C allele ( Table 7 ). Given the estimated fitness levels and assuming that the fitness level of AA , AC and CC are w AA , w AC and w CC , respectively, then
Using the data in Table 7 , the estimates are of s =0.166 and of h =0.518. In other words, it appears that the advantageous effect of the C allele is very close to additivity ( h =0.5) and is not recessive in its advantageous effect. Interestingly, if the C allele is more recessive, that is, the fitness of AC is lower, then the C allele cannot invade a population in which the S allele is at equilibrium.
S and α-thalassemia
Two hemoglobinopathies, sickle cell and α + -thalassemia, occur in high frequency in sub-Saharan Africa. Williams et al. (2005a) examined these disorders in Kenya in a population that was segregating for both variants. In this population, they found that the protection from malaria given by each variant was lost when the two disorders were inherited together in the same individual (see also May et al., 2007 ). They suggested that this negative epistasis could be the reason why α + -thalassemia has not been fixed in any population in sub-Saharan Africa. This situation has been examined theoretically by Williams et al. (2005a) and Penman et al. (2009) , using an epidemiological approach. Here, I will use a traditional population genetics approach and show that the results are generally consistent with their findings, using an epidemiological model. An important caveat is that although most general properties discussed here are enlightening and significant, some conclusions can depend on rather small differences in estimated values ( Williams et al., 2005a ; Penman et al., 2009 ; Hedrick, 2011b ).
As background for discussion of the fitness array for these two loci, let us examine what the expectation is when there are independent effects on viability for the two loci (no epistasis on a multiplicative scale Hedrick, 2011a , p.554). Table 8 lists on the left margin, the relative fitness values for the β-globin genotypes AA , AS and SS of 1– s , 1 and 0, respectively, and on the top, the relative fitness values for α-globin genotypes αα/αα, −α/αα and −α/−α of 1, 1+ ht and 1+ t , respectively. The product of these marginal values yields the expected fitness levels in the table.
The expected relative viabilities for two-locus genotypes without epistasis can be calculated in the following manner. First, the difference in the relative values for genotypes AS αα/αα and AA αα/αα yields an estimate of s . The fitness of genotype AA αα/αα divided by the fitness of genotype AA −α/−α yields an estimate of 1+ t . Therefore, for the example in Table 9b that I will discuss below, s =0.448 and t =0.275, resulting in a value of 1.275 for the expected fitness without epistasis of genotype AS −α/−α. The estimated value for this genotype of 0.554 is substantially lower than this expectation and illustrates negative epistasis.
To calculate the relative viabilities of the different two-locus genotypes (the approach used here was suggested by B. Penman), genotype-specific annual mortality rates were used based on the relative rates of hospital admissions in children with severe malaria ( Table 9a ; from legend of Figure 2 in Williams et al. (2005a) ). To each of these mortality rates, an annual non-genotype specific mortality rate was added (in the example here, a rate of 0.034 was added) to yield a total annual mortality rate for each genotype. The reciprocal of these annual mortality rates provides an estimate of the relative life expectancies. When these values are standardized by the largest relative life expectancy (for these data the genotype AS αα/αα has the maximum value of 29.41), the relative viability values in Table 9b are obtained.
When there is a wild-type genotype at both loci, the estimated relative survival is only 0.552. Notice that both the presence of S in AS and the presence of −α in both genotypes −α/αα and −α/−α increases the estimated relative survival, given normal genotypes at the other locus. However, when there is an AS genotype and homozygosity for −α, the relative survival is reduced to only 0.554, essentially the same relative survival for the wild-type genotype.
There are several things to note about this array of relative viability values. First, when only the normal αα haplotype at the α-globin gene is present (first column), there is a stable polymorphism with S having a frequency of 0.309. Second, when only the normal A allele is present at the β-globin locus (first row), the −α/−α genotype has the highest fitness, predicting that this variant haplotype would be fixed (unless there is a negative effect of anemia). Overall then, the normal AA αα/αα genotype has a low relative viability in this malarial environment.
We can intuit some two-locus behavior by examining these viability values further. For example, if the population is polymorphic for A and S , it appears that −α will be able to increase from a low frequency in the population (enter) because the relative viability of AA −α/αα is greater than that of AA αα/αα (the most frequent genotype at this point; first row of Table 9b ) and AS −α/αα is only slightly less viable than AS αα/αα (second row). On the other hand, if the population is fixed for −α−α, it does not appear that S can enter the population because AA −α/−α > AS −α/−α (last column of Table 9b ).
However, the genetic behavior of this two-locus fitness array has some further interesting properties that are observed if the two-locus gamete frequency equations with selection ( Hedrick, 2011a , p. 559) are iterated. First, if the population is polymorphic at the equilibrium for A and S and −α haplotype enters the population, the system will increase to a two-locus stable equilibrium with −α having a frequency of 0.272 and S having a frequency of 0.256, with no linkage disequilibrium. Second, if the −α is polymorphic, there is an unstable equilibrium such that −α will, depending upon its initial frequency, either continue to increase or go to the two-locus equilibrium discussed above, if S is introduced into the population. Figure 3 presents this situation in which the initial frequency of S is 0.01 and the initial frequency of −α is either 0.55 or 0.65. For 0.55, the frequency of −α initially increases and then approaches the two-locus equilibrium at 0.272. On the other hand, when the initial frequency is 0.65, it continues to increase toward unity whereas S initially increases but then decreases to 0. In other words, the presence of S maintains the frequency of −α lower when the fitness of AS −α/−α is low (high negative epistasis). If fitness of AS −α/−α is somewhat higher (less negative epistasis), then −α can be fixed for all initial frequencies.
The change in the frequency of S where the initial frequency is 0.01 and the change in the frequency of −α where the initial frequency is either 0.55 or 0.65 using the relative fitness levels in Table 9b .
Conclusions
Genetic resistance to malaria in humans provides a rich source of data and examples of many aspects of population genetics, particularly cases of strong selection. For example, maintenance of the sickle-cell hemoglobin variant is the classic example of heterozygote advantage, G6PD deficiency is an example of very strong selection at an X-linked locus, variants at the β-globin gene ( S , C and E ) and causing G6PD deficiency ( A -, Med and Mahidol ) illustrate selective advantage from a single-nucleotide change, HLA-B53 provides an example of a selective advantage of a gene conversion product and α-thalassemia provides an example of selection on duplicated loci. In addition, understanding the geographic patterns of these variants includes consideration of selection as well as gene flow, mutation, genetic drift and the interaction of these factors. For example, to illustrate the importance of gene flow, most Mediterranean S alleles appear to descend from the African Benin haplotype ( Flint et al., 1998 ) and the Southeast Asian deletion of both α- globin genes, −− SEA , is widespread in a number of Asian populations ( Chui and Waye 1998 ; Bernini 2001 ). Estimating the age during which these variants have been strongly selected also includes incorporating information about recombination in the genomic region surrounding the selected variants.
An aspect of population genetics that will probably become even more important is the joint impact of multiple malaria resistance variants. Here, I discuss future changes in populations segregating for the two β-globin variants S and C and predict that, given the continued presence of malaria, selection will result in the elimination of S and fixation of C . In populations segregating for α-thalassemia and S , variants at two different loci, because of negative epistasis, either the α-thalassemia haplotype is predicted to be fixed and the S allele eliminated or a stable equilibrium of both variants is predicted depending upon the starting frequencies. Although these results are initially somewhat counterintuitive, one needs to realize that they are dependent upon specific estimated selection coefficients with large confidence intervals and which may vary over time (in the past or in the future) and may vary in different populations.
Obviously, Haldane (1949b) was correct to point out the potential importance of genetic variants in humans for resistance to malaria. Genes conferring resistance to malaria provide some of the best-known case studies of strong positive selection in modern humans and the sickle-cell hemoglobin variant remains the classic example of heterozygote advantage. Overall, the original ‘malaria hypothesis’ of Haldane that diseases such as thalassemia are polymorphisms with an advantage to heterozygotes in malarial environments has been proven correct; however, much is still to be learned about actual mechanisms of protection, other genes that confer resistance and the population genetics of this variation.
Ackerman H, Usen S, Jallow M, Sisay-Joof F, Pinder M, Kwiatkowski DP (2005). A comparison of case-control and family-based association methods: the example of sickle-cell and malaria. Ann Hum Genet 69 : 559–565.
Google Scholar
Agarwal A, Guindo A, Cissoko Y, Taylor JG, Coulibaly D et al . (2000). Hemoglobin C associated with protection from severe malaria in the Dogon of Mali, a West African population with a low prevalence of hemoglobin S. Blood 96 : 2358–2363.
Agresti A (2007). An Introduction to Categorical Data Analysis , 2nd ed. John Wiley & Sons: New York.
Allen SJ, O'Donnell A, Alexander ND, Alpers MP, Peto TE et al . (1997). Alpha+thalassemia protects children against disease caused by other infections as well as malaria. Proc Nat Acad Sci USA 94 : 14736–14741.
Allen SJ, O'Donnell A, Alexander NDE, Mgone CS, Peto TEA et al . (1999). Prevention of cerebral malaria in children in Papua New Guinea by Southeast Asian ovalocytosis band 3. Am J Trop Med Hyg 60 : 1056–1060.
Allison AC (1954). Protection afforded by the sickle-cell trait against subtertian malaria infection. Br Med J 1 : 290–294.
Allison AC (1955). Aspects of polymorphism in man. Cold Spring Harbor Symp Quant Biol 20 : 239–251.
Allison AC (1956). The sickle-cell and haemoglobin C genes in some African populations. Ann Hum Genet 21 : 67–89.
Allison AC (1964). Polymorphism and natural selection in human populations. Aspects of polymorphism in man. Cold Spring Harbor Symp Quant Biol 29 : 137–149.
Allison AC (2004). Two lessons from the interface of genetics and medicine. Genetics 166 : 1591–1599.
Allsopp CEM, Hill AVS, Kwiatkowski D, Hughes A, Bunce M et al . (1991). Sequence analysis of HLA-Bw53, a common west African allele, suggests and origin by gene conversion of HLA-B53. Hum Immunol 30 : 105–109.
Ayodo G, Price AL, Keinon A, Ajwang A, Otieno MF et al . (2007). Combining evidence of natural selection with association analysis increases power to detect malaria-resistance variants. Am J Hum Genet 81 : 234–242.
Beet EA (1946). Sickle cell disease in the Balovale district in Northern Rhodesia. East Afr Med J 23 : 75–86.
Bernini LK (2001). Geographic distribution of α thalassemia. In: Steinberg MH, Forget BG, Higgs DR, Nagel RL (eds). Disorders of Hemoglobin: Genetics, Pathophysiology and Clinical Management . Cambridge University Press: Cambridge. pp 878–894.
Bonham VL, Dover GJ, Brody LC (2010). Screening student athletes for sickle cell trait-a social and clinical experiment. New Eng J Med 363 : 997–999.
Calafell F, Roubinet F, Ramirez-Soriano A, Saitou N Bertranpetit J, Blancher A (2008). Evolutionary dynamics of the human ABO gene. Hum Genet 124 : 123–135.
Cappellini MD, Fiorelli G (2008). Glucose-6-phosphate dehydrogenase deficiency. Lancet 371 : 64–74.
Carter R, Mendis KN (2002). Evolutionary and historical aspects of the burden of malaria. Clin Microbiol Rev 15 : 564–594.
Cattani JA, Gibon FD, Alpers MP, Crane GG (1987). Hereditary ovalocytosis and reduced susceptibility to malaria in Papua New Guinea. Trans Roy Soc Trop Med Hyg 81 : 705–709.
Chotivanich K, Udomsangpetch R, Pattanapanyasat K, Chierakul W, Simpson J et al . (2002). Hemoglobin E: a balanced polymorphism protective against high parasitemias and thus severe P . falciparum malaria Blood 100 : 1172–1176.
Chui DHK, Waye JS (1998). Hydrops felatis caused by α-thalassemia: an emerging health care problem. Blood 91 : 2213–2222.
Clark TG, Diakite M, Auburn S, Campino S, Fry AE et al . (2009). Tumor necrosis factor and lymphotoxin-α polymorphisms and severe malaria in African populations. J Infect Dis 199 : 569–575.
Coluzzi M, Sabatini A, Torre A, Di Deco MA, Petrarca V. (2002). A polytene chromosome analysis of the Anopheles gambiae species complex. Science 298 : 1415–1418.
Connes P, Reid H, Hardy-Dessources M-D, Morrison E, Hue O (2008). Physiological responses of sickle cell trait carriers during exercise. Sports Med 38 : 931–946.
Cox-Singh J, Davis TME, Lee K-S, Shamsul SSG, Matusop A (2008). Plasmodium knowlesi malaria in human is widely distributed and potentially life threatening. Clin Inf Dis 46 : 165–171.
Crow JF (2004). J B. S. Haldane's ideas in biology with special reference to disease and evolution. In: Dronamraju KR (ed). Infectious Disease and Host-Pathogen Evolution . Cambridge University Press: Cambridge. pp 11–17.
Cserti CM, Dzik WH (2007). The ABO blood group system and Plasmodium falciparum malaria. Blood 110 : 2250–2258.
Currat M, Trabucher G, Rees D, Perrin P, Harding RM et al . (2002). Molecular analysis of the β-globin gene cluster in the Niokholo Mandenka population reveals a recent origin of the β S Senegal mutation. Am J Hum Genet 70 : 207–223.
Curtin PD (1989). Death by Immigration: Europe's Encounter with the Tropics in the Nineteenth Century . Cambridge Univ Press: New York.
Davenport MP, Quinn CL, Chicz RM, Green BN, Willis AC et al . (1995). Naturally processed peptides from two disease-resistance associated HLA-DR13 alleles show related sequence motifs and the effects of the dimorphism at position 86 of the HLA DRβ chain. Proc Nat Acad Sci USA 92 : 6567–6571.
Dolo A, Modiano D, Maiga B, Daou M, Dolo G et al . (2005). Difference in susceptibility to malaria between two sympatric ethnic groups in Mali. Am J Trop Med Hyg 72 : 243–248.
Dronamraju KR (ed) (2004). Infectious Disease and Host-Pathogen Evolution . Cambridge University Press: Cambridge.
Eid NA, Hussein AA, Elzein AM, Mohamed HS, Rockett K et al . (2010). Candidate malaria susceptibility/protective SNPs in hospital and population-based studies: the effect of sub-structuring. Malaria J 9 : 119.
Ferwerda B, McCall MBB, Alonso S, Giamarellos-Bourboulis EJ, Mouktaroudi M et al . (2007). TLR4 polymorphisms, infectious diseases, and evolutionary pressure during migration of modern humans. Proc Nat Acad Sci USA 42 : 16645–16650.
Flint J, Harding RM, Boyce AJ, Clegg JB (1998). The population genetics of the haemoglobinopathies. Bailliere Clin Haematol 11 : 1–51.
Flint J, Hill AV, Bowden DK, Oppenheimer SJ, Sill PR et al . (1986). High frequencies of alpha-thalassemia are the result of natural selection by malaria. Nature 321 : 744–750.
Fry AE, Ghansa A, Small KS, Palma A, Auburn S et al . (2009). Positive selection of a CD36 nonsense variant in sub-Saharan Africa, but no association with severe malaria phenotypes. Hum Molec Genet 18 : 2683–2693.
Fry AE, Griffiths MJ, Auburn S, Diakite M, Forton JT et al . (2008). Common variation in the ABO glycosyltransferase is associated with susceptibility to severe Plasmodium falciparum malaria. Hum Molec Genet 17 : 567–576.
Fucharoen S (2001). Hemoglobin E disorders. In: Steinberg MH, Forget BG, Higgs DR, Nagel RL (eds). Disorders of Hemoglobin: Genetics, Pathophysiology and Clinical Management . Cambridge: Cambridge Univ. Press:Cambridge. pp. 1139–1154.
Fucharoen G, Fucharoen S, Sanchaisuriya K, Sae-ung N, Suyasunanond U et al . (2002). Frequency distribution and haplotypic heterogeneity of β E -globin gene among eight minority groups of northeast Thailand. Hum Hered 53 : 18–22.
Garrigan D, Hedrick PW (2003). Detecting adaptive molecular evolution, lessons from the MHC. Evolution 57 : 1707–1722.
Genton B, Al-Yaman F, Mgone CS, Alexander N, Paniu MM, Aplers MP (1995). Ovalocytosis and cerebral malaria. Nature 378 : 564–565.
Ghosh K (2008). Evolution and selection of human leukocyte antigen alleles by Plasmodium falciparum infection. Hum Immunol 69 : 856–860.
Giardine B, van Baal S, Kaimakis P, Riemer C, Miller W et al . (2007). HbVar database of human hemoglobin variants and thalassemia mutations: 2007 update. Hum Mut 28 : 206.
Gouagna LC, Bancone G, Yao F, Yameogo B, Dabiré KR et al . (2010). Genetic variation in human HBB is associated with Plasmodium falciparum transmission. Nat Genet 42 : 328–331.
Guerra CA, Howes RE, Patil AP, Gething PW, Van Boeckel TP et al . (2010). The international limits and population at risk of Plasmodium vivax transmission in 2009. PLoS Neglected Trop Dis 4 : e774.
Guindo A, Fairhurst RM, Doumbo OK, Wellems TE Diallo DA (2007). X-linked G6PD deficiency protects hemizygous males but not heterozygous females against severe malaria. PLoS Med 4 : e66.
Haldane JBS (1949a). Disease and evolution. Ric Sci Suppl 19 : 68–76.
Haldane JBS (1949b). The rate of mutation of human genes. Hereditas 35 (S1): 267–273.
Hamblin MT, Di Rienzo A (2000). Detection of the signature of natural selection in humans: evidence from the Duffy blood group locus. Am J Hum Genet 66 : 1669–1679.
Hamblin MT, Thompson EE, Di Rienzo A (2002). Complex signatures of natural selection at the Duffy blood group locus. Am J Hum Genet 70 : 369–383.
Hanchard N, Elzein A, Trafford C, Rockett K, Pinder M. et al . (2007). Classical sickle beta-globin haplotypes exhibit a high degree of long-range haplotype similarity in African and Afro-Caribbean populations. BMC Genet 8 : 52.
Hedrick PW (2004). Estimation of relative fitnesses from relative risk data and the predicted future of haemoglobin alleles S and C . J Evol Biol 17 : 221–224.
Hedrick PW (2011a). Genetics of Populations . 4th ed. Jones and Bartlett: Boston.
Hedrick PW (2011b). Selection and mutation for α thalassemia in non-malarial and malarial environments. Ann Hum Genet (in press).
Hedrick PW, Kim T (2000). Genetics of complex polymorphisms: parasites and maintenance of MHC variation. In: Singh R. Krimbas C (eds). Evolutionary Genetics: From Molecules to Morphology . Cambridge University Press: Cambridge. pp 204–234.
Hill AVS (1991). HLA associations with malaria in Africa: some implications for MHC evolution. In: Klein J, Klein D (eds). Molecular Evolution of the Major Histocompatibility Complex . Springer-Verlag, Berlin. pp 403–434.
Hill AVS, Allsopp CEM, Kwiatkowski D, Anstey NM, Twumasi P et al . (1991). Common west African HLA antigens are associated with protection from severe malaria. Nature 352 : 595–600.
Hill AVS, Bowden DK, O'Shaughnessy DF, Weatherall DJ, Clegg JB (1988). β thalassemia in Melanesia: association with malaria and characterization of a common variant (IVS1 nt 5 G-C). Blood 72 : 9–14.
Hill AVS, Yates SNR, Allsopp CEM, Gupta A, Gilbert SC et al . (1994). Human leukocyte antigens and natural selection by malaria. Phil Trans Roy Soc Lond B 346 : 379–385.
Ingram VM (2004). Sickle-cell anemia hemoglobin: the molecular biology of the first ‘molecular disease’- the crucial importance of serendipity. Genetics 167 : 1–7.
Jallow M, Teo YY, Small KS, Rockett KA, Deloukas P et al . (2009). Genome-wide and fine-resolution association analysis of malaria in West Africa. Nat Genet 41 : 657–665.
Jarolim P, Palek J, Amato D, Hassan K, Sapak P, Nurse GT et al . (1991). Deletion in erythrocyte band 3 gene in malaria–resistant Southeast Asian ovalocytosis. Proc Nat Acad Sci USA 88 : 11022–11026.
Johnson MK, Clark TD, Njama-Meya D, Rosenthal PJ, Parik (2009). Impact of the method of G6PD deficiency assessment on genetic association studies of malaria susceptibility. PLoS ONE 4 : 9 (e7246).
Joy DA, Feng X, Mu J, Furuya T, Chotivanich K et al . (2003). Early origin and recent expansion of Plasmodium falciparum . Science 300 : 318–321.
Karasov T, Messer PW, Petrov DA (2010). Evidence that adaptation in Drosophila is not limited by mutation at single sites. PLoS Genet 6 (6): e1000924.
Kasehagen LJ, Mueller I Kiniboro B, Bockarie MJ, Reeder JC et al . (2007). Reduced Plasmodium vivax erythrocyte infection in PNG Duffy-negative heterozygotes. PLoS ONE 2 (3): e336.
Kaslow RA, McNicholl JM, Hill AVS (2008). Genetic Susceptibility to Infectious Diseases . Oxford University Press: Oxford.
Kermarrec N, Roubinet F, Apoil PA, Blancher A (1999). Comparison of allele O sequences of the human and non-human primate ABO system. Immunogenetics 49 : 517–526.
Kimura M, Soemantri A, Ishida T (2002). Malaria species and Southeast Asian ovalocytosis defined by a 27-bp deletion in the erythrocyte band 3 gene. Southeast Asian J Trop Med Pub Health 33 : 4–6.
Kwiatkowski DP (2005). How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet 77 : 171–192.
Kwiatkowski DP, Luoni G (2008). Malaria. In: Kaslow RA, McNicholl JM, Hill AVS (eds). Genetic Susceptibility to infections Diseases . Oxford University Press. pp 372–386.
Lam K-WG, Jeffreys AJ (2006). Processes of copy-number change in human DNA: the dynamics of α-globin gene deletion. Proc Nat Acad Sci USA 103 : 8921–8927.
Langhorne J, Ndungu FM, Sponaas A-M, Marsh K (2008). Immunity to malaria: more questions than answers. Nat Immunol 9 : 723–732.
Lawniczak MKN, Emrich SJ, Holloway AK, Regier AP, Olson M et al . (2010). Widespread divergence between incipient Anophoeles gambiae species revealed by whole genome sequence. Science 330 : 512–514.
Leslie T, Briceño, Mayan I, Mohammed N, Klinkenberg E et al . (2010). The impact of phenotypic and genotypic G6PD deficiency on risk of Plasmodium vivax infection: A case-control study amongst Afghan refugees in Pakistan. PLoS Med 7 : e1000283.
Liu W, Li V, Rudicell RS, Robertson JD, Keele BF et al . (2010). Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature 467 : 420–425.
Livingstone FB (1958). Anthropological implications of sickle cell gene distribution in west Africa. Am Anthrop 60 : 533–562.
Livingstone FB (1967). Abnormal Hemoglobins in Human Populations . Aldine: Chicago.
Livingstone FB (1985). Frequencies of Haemoglobin Variants . Oxford University Press: Oxford.
López C, Saravia C, Gomez A, Hoebeke J, Patarroyo MA (2010). Mechanisms of genetically-based resistance to malaria. Gene 467 : 1–12.
Louicharoen C, Patin E, Paul R, Nuchprayoon I, Witoonpanich B et al . (2009). Positively selected G6PD-Mahidol mutation reduces Plasmodium vivax density in southeast Asian. Science 326 : 1546–1549.
MacFie TS, Nerrienet E, Bontrop RE, Mundy NI (2009). The action of falciparum malaria on the human and chimpanzee genomes compared: absence of evidence for a genomic signature of malaria at HBB and G6PD in three subspecies of chimpanzee. Infec Genet Evol 9 : 1248–1252.
Mackinnon MJ, Mwangi TW, Snow RW, Marsh K, Williams TN (2005). Heritability of malaria in Africa. PLoS Med 2 : 1253–1259. (e340).
Mason PJ, Bautista JM, Gilsanz F (2007). G6PD deficiency: the genotype-phenotype association. Blood Rev 21 : 267–283.
May J, Evans JA, Timmann C, Ehmen C, Busch W et al . (2007). Hemoglobin variants and disease manifestations in severe falciparum malaria. J Am Med Assoc 297 : 2220–2226.
Ménard D, Barnakas C, Bouchier C, Henry-Halldin C, Gray LR et al . (2010). Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Nat Acad Sci USA 107 : 5967–5971.
Miller LH, Mason SJ, Clyde DF, McGinnis MH (1976). The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype FyFy. New Engl J Med 295 : 302–304.
Mockenhaupt FP, Ehrhardt S, Gellert S, Otchwemah RN, Dietz E et al . (2004). Alpha (+)- thalassemia protects African children from severe malaria. Blood 104 : 2003–2006.
Modiano G, Morpurgo G, Terrenato L, Novelletto A, Di Rienzo A et al . (1991). Protection against malaria morbidity: near-fixation of the α-thalassemia gene in a Nepalese population. Am J Hum Genet 48 : 390–397.
Modiano D, Luoni G, Sirima BS, Simpore J, Verra F et al . (2001). Haemoglobin C protects against clinical Plasmodium falciparum malaria. Nature 414 : 305–308.
Modiano D, Bancone G, Ciminelli BM, Pompei F, Blot I (2008). Haemoglobin S and haemoglobin C: ‘quick but costly’ versus ‘slow but gratis’ genetic adaptations to Plasmodium falciparum malaria. Hum Molec Genet 17 : 789–799.
Mueller I, Widmer S, Michel D, Maraga S, McNamara DT et al . (2009). High sensitivity detection of Plasmodium species reveals positive correlations between infections of different species, shifts in age distribution and reduced local variation in Papua New Guinea. Malaria J 8 : 41.
Neafsey DE, Lawniczak MKN, Park DJ, Redmond SN, Coulibaly MB et al . (2010). SNP genotyping defines complex gene-flow boundaries among African malaria vector mosquitoes. Science 330 : 514–517.
Neel JV, Valentine WN (1947). Further studies on the genetics of thalassemia. Genetics 32 : 38–63.
Nkhoma ET, Poole C, Vannappagari V, Hall SA, Beutler E (2009). The global prevalence of glucose-6-phosphate dehydrogenase deficiency: A systematic review and meta-analysis. Blood Cells Molec Dis 42 : 267–278.
O'Donnell A, Premawardhena A, Arambepola M, Samaranayake R, Allen SJ et al . (2009). Interaction of malaria with a common form of severe thalassemia in an Asian population. Proc Nat Acad Sci USA 106 : 18716–18721.
Ohashi J, Naka I, Patarapotikul J, Hananantachai H, Brittenham G. et al . (2004). Extended linkage disequilibrium surrounding the hemoglobin E variant due to malarial selection. Am J Hum Genet 74 : 1198–1208.
Ohashi J, Naka I, Patarapotikul J, Hananantachai H, Brittenham G. et al . (2005). Strong linkage disequilibrium of a HbE variant with the (AT)9(T)5 repeat in the BP1 binding site upstream of the β-globin gene in the Thai population. J Hum Genet 50 : 7–11.
O'Shaughnessy DF, Hill AVS, Bowden DK, Weatherall DJ, Clegg JB (1990). Globin genes in Micronesia: origins and affinities of Pacific Island peoples. Am J Hum Genet 46 : 144–155.
Pauling L, Itano HA, Singer SJ, Wells IC (1949). Sickle cell anemia a molecular disease. Science 110 : 543–548.
Penman BS, Pybus OG, Weatherall DJ, Gupta S (2009). Epistatic interactions between genetic disorders of hemoglobin can explain why the sickle-cell gene is uncommon in the Mediterranean. Proc Nat Acad Sci USA 106 : 21242–21246.
Platt OS, Brambilla DJ, Rosse WF, Milner PF, Castro O et al . (1994). Mortality in sickle cell disease. New Eng J Med 330 : 1639–1644.
Ralph P, Coop G (2010). Parallel adaptation: one or many waves of advance of an advantageous allele? Genetics 186 : 647–668.
Rowe AK, Rowe SY, Snow RW, Korenromp EL, Schellenberg JR et al . (2006). The burden of malaria mortality among African children in the year 2000. Int J Epidemiol 35 : 691–704.
Rowe JA, Handel IG, Thera MA, Deans A-M, Lyke KE et al . (2007). Blood group O protects against severe Plasmodium falciparum malaria through the mechanism of reduced rosetting. Proc Nat Acad Sci USA 104 : 17471–17476.
Ruwende C, Khoo SC, Snow RW, Yates SNR, Kwiatkowski D et al . (1995). Natural selection of hemi- and heterozygotes for G6PD deficiency in Africa by resistance to severe malaria. Nature 376 : 246–249.
Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ et al . (2002). Detecting recent positive selection in the human genome from haplotype structure. Nature 419 : 832–837.
Sanchaisuriya K, Fucharoen G, Sae-ung N, Siriratmanawong N, Surapot S, Fucharoen S. (2001). Molecular characterization of hemoglobin C in Thailand. Am J Hematol 67 : 189–193.
Saunders MA, Hammer MF, Nachman MW (2002). Nucleotide variability at G6pd and the signature of malarial selection in humans. Genetics 162 : 1849–1861.
Seixas S, Ferrand N, Rocha J (2002). Microsatellite variation and evolution of the human Duffy blood group polymorphism. Molec Biol Evol 19 : 1802–1806.
Sheng A-M, Chertow DS, Morens DM, Taubenberger JK (2010). Fatal 1918 pneumonia case complicated by erythrocyte sickling. Emerg Infect Dis 16 : 2000–2001.
Siniscalco M, Bernini L, Latte B, Motulsky AG (1961). Favism and thalassemia in Sardinia and their relationship to malaria. Nature 190 : 1179–1180.
Slatkin M (2008). A Bayesian method for jointly estimating allele age and selection intensity. Genet Res 90 : 129–137.
The Malaria Genomic Epidemiology Network (2008). A global network for investigation the genomic epidemiology of malaria. Nature 456 : 732–737.
Timmann C, Evan JA, König IR, Kleensang A Rüschendorf F et al . (2007). Genome-wide linkage analysis of malaria infection intensity and mild disease. PLoS Genet 3 : e48.
Tishkoff SA, Varkonyl R, Cahinhinan N, Abbes S, Argyropoulos G et al . (2001). Haplotype diversity and linkage disequilibrium at human G6PD : recent origin of alleles that confer malarial resistance. Science 293 : 455–45. 462.
Tishkoff SA, Verrelli BC (2004). G6PD deficiency and malarial resistance in humans: insight from evolutionary genetic analysis. In: Dronamraju KR (ed). Infectious Disease and Host-Pathogen Evolution . Cambridge University Press: Cambridge. pp 113–140.
Tsaras G, Owusu-Ansah A, Boateng FO, Amoateng-Adjepong Y (2009). Complications associated with sickle cell trait: a brief narrative review. Amer J Med 122 : 507–512.
Tung J, Primau A, Bouley AJ, Severson TF, Alberts SC, Wray GA (2009). Evolution of a malaria resistance gene in wild primates. Nature 460 : 388–391.
Uneke CJ (2007). Plasmodium falciparum malaria and ABO blood group: is there any relationship. Parasitol Res 100 : 769–765.
Verra F, Mangang VD, Modiano D (2009). Genetics of susceptibility to Plasmodium flaciparum: from classical malaria resistance genes towards genome-wide association studies. Parasite Immunol 31 : 234–253.
Verrelli BC, McDonald JH, Argyropoulos G, Destro-Bisol G, Froment A et al . (2002). Evidence for balancing selection from nucleotide sequence analyses of human G6PD . Am J Hum Genet 71 : 1112–1128.
Verrelli BC, Tishkoff SA, Stone AC, Touchman JW (2006). Contrasting histories of G6PD molecular evolution and malarial resistance in humans and chimpanzees. Mol Biol Evol 23 : 1592–1601.
Weatherall DJ (2004). J. B. S. Haldane and the malaria hypothesis. In: Dronamraju KR (ed). Infectious Disease and Host-Pathogen Evolution . Cambridge University Press. pp 18–36.
Weatherall DJ (2008). Genetic variation and susceptibility to infection: the red cell and malaria. Br J Haematol 141 : 276–286.
Weatherall DJ (2010). The inherited diseases of hemoglobin are an emerging global health burden. Blood 115 : 4331–4336.
Weatherall DJ, Clegg JB (2001). The Thalassemia Syndromes , 4th ed. Blackwell Scientific, Oxford.
WHO (1998). Demographic data for health situation assessment and projections . WHO HST/HSP: Geneva.
WHO (2009). World Malaria Report . World Health Organization, Geneva.
Wierenga KJJ, Hambleton IR, Lewis NA (2001). Survival estimates for patients with homozygous sickle-cell disease in Jamaica: a clinic-based population study. Lancet 357 : 680–683.
Williams TN, Mwangi TW, Wambua S, Peto TEA, Weatherall DJ. (2005a). Negative epistasis between the malaria-protective effects of α + -thalassemia and the sickle cell trait. Nat Genet 37 : 1253–1262.
Williams TN, Wambua S, Uyoga S, Macharia A, Mwacharo JK et al . (2005b). Both heterozygous and homozygous α + thalassemias protect against severe and fatal Plasmodium falciparum malaria on the coast of Kenya. Blood 106 : 368–371.
Willcox M, Björkman A, Brohult J (1983). Falciparum malaria and β-thalassemia trait in northern Liberia. Ann Trop Med Parasit 77 : 335–347.
Wood ET, Stover DA, Slatkin M, Nachman MW, Hammer MF 2005)). The β-globin recombinational hotspot reduces the effects of strong selection around HbC, a recently arisen mutation providing resistance to malaria. Am J Hum Genet 77 : 637–642.
Yamamoto F, Clausen H, White T, Marker J, Hakomori S (1990). Molecular genetic basis of the histo-blood group ABO system. Nature 345 : 229–233.
Zimmerman PA (2004). The enigma of Plasmodium vivax malaria and erythrocyte Duffy negativity. In: Dronamraju KR (ed.). Infectious Disease and Host-pathogen Evolution . Cambridge University Press: Cambridge. pp 141–172.
Zimmerman PA, Woolley I, Masinde GL, Miller SM, McNamara D et al . (1999). Emergence of FY* null in a Plasmodium vivax -endemic region of Papua New Guinea. Proc Nat Acad Sci USA 96 : 13973–13977.
Download references
Acknowledgements
I appreciate the comments on various aspects of the manuscript by B Penman, M Slatkin, A Stone, J Taylor, G Thomson, B Verrelli, P Zimmerman and three anonymous reviewers, and the assistance of M Saraniti on the Italian translation.
Author information
Authors and affiliations.
School of Life Sciences, Arizona State University, Tempe, AZ, USA
P W Hedrick
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to P W Hedrick .
Ethics declarations
Competing interests.
The author declares no conflict of interest.
Rights and permissions
Reprints and permissions
About this article
Cite this article.
Hedrick, P. Population genetics of malaria resistance in humans. Heredity 107 , 283–304 (2011). https://doi.org/10.1038/hdy.2011.16
Download citation
Received : 29 November 2010
Revised : 07 February 2011
Accepted : 11 February 2011
Published : 23 March 2011
Issue Date : October 2011
DOI : https://doi.org/10.1038/hdy.2011.16
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- age of allele
- sickle cell
- thalassemia
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies

IMAGES